Description
Mechanistic target of rapamycin (mTOR), a serine/threonine protein kinase, regulates biological processes in response to changes in nutrients or metabolites (Reviewed in Liu and Sabatini, 2020). Inhibition of mTOR signaling delays aging and increases lifespan in multiple organisms by downregulating substrates, including S6 kinase (S6K) that phosphorylates ribosomal subunit 6 (Reviewed in Kennedy and Lamming, 2016). Genetic inhibition of RSKS-1, the Caenorhabditis elegans S6 kinase, extends lifespan via decreasing protein synthesis and upregulating longevity-promoting factors (Reviewed in Johnson et al., 2013; Lee et al. 2015). However, the longevity phenotypes of previously characterized loss-of-function rsks-1 mutants are variable and greatly affected by environmental factors, such as temperatures (Seo et al., 2013; Miller et al., 2017). Here, we aimed at identifying rsks-1 mutants whose lifespan phenotypes are robust and reproducible at 20°C, the standard temperature for C. elegans lifespan assays.
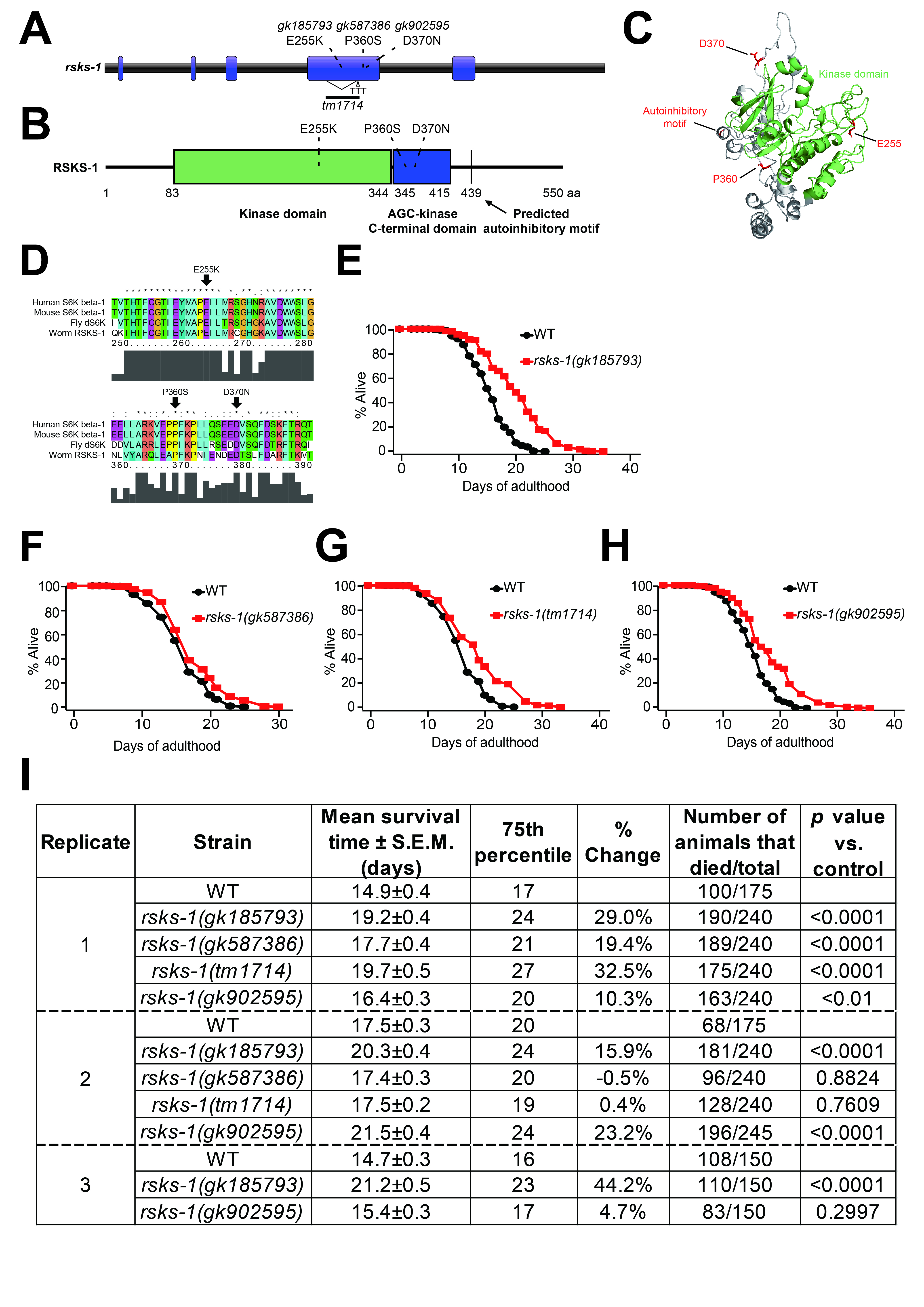
We measured the lifespan of three available rsks-1 missense mutants generated by the Million Mutant Project (Thompson et al., 2013) after outcrossing four times to wild-type (N2) strains, without treatment with 2′-deoxy-5-fluorouridine. The three missense mutant alleles are rsks-1(gk185793), rsks-1(gk587386), and rsks-1(gk902595), which respectively cause E255 to K, P360 to S, and D370 to N changes (Fig. 1A–D). We also measured the lifespan of previously characterized rsks-1(tm1714) deletion mutants (Fig. 1A) (Chen et al., 2013; Seo et al., 2013). We found that rsks-1(gk185793), which encodes RSKS-1 E255K, caused a consistent and strong longevity phenotype in all three independent trials (Fig. 1E and I). In contrast, rsks-1(gk587386) and rsks-1(tm1714) mutations extended lifespan in one out of two trials (Fig. 1F, G, and I). We found that rsks-1(gk902595) lengthened lifespan slightly (p<0.01) or robustly (p<0.0001), or did not (p=0.2997) in three trials (Fig. 1H and I). These variations in the lifespan of the three latter rsks-1 mutants (Fig. 1F–I) may have been caused by small changes in environmental factors such as temperatures that affect the lifespan of rsks-1 mutants (Seo et al., 2013; Miller et al., 2017). Together, our data indicate that rsks-1(gk185793) mutant allele that alters the acidic E255 residue to a basic K increases the lifespan of C. elegans reproducibly and robustly.
We speculate that the effects of rsks-1(gk185793) on lifespan were more robust than other missense alleles tested, because of following two reasons. First, E255 resides in the catalytic kinase domain of RSKS-1/S6K, but the others are located in the AGC-kinase C-terminal domain (Fig. 1A–D). Second, E to K, a change from an acidic to a basic amino acid, is likely to cause a bigger impact on the protein function than P to S and D to N changes. Future studies with additional missense mutations will clarify this issue.
Our study suggests that rsks-1(gk185793) mutant C. elegans is an excellent tool for aging research as a reliable genetic model for longevity conferred by reduced mTOR signaling. Additional future studies will enhance the value of rsks-1(gk185793) mutants for research on aging and mTOR signaling. First, further outcross of rsks-1(gk185793) mutants to wild-type worms to remove remaining background mutations will corroborate the causality of change in RSKS-1 for longevity. Alternatively or additionally, generation of the identical missense mutant allele by using CRISPR genome editing will be helpful. Second, confirming whether rsks-1(gk185793) is a reduction-of-function allele will be crucial for establishing the rsks-1 mutant allele as a bona fide genetic model for reduced mTOR signaling. To this end, the assessment of phosphorylated ribosomal S6 and global protein synthesis levels will be required. Third, performing lifespan assays at various temperatures (e.g. 15°C and 25°C) and on different diets (e.g. E. coli HT115 and HB101 strains) may extend the utility of rsks-1(gk185793) mutants as a general longevity model. Lastly, E255 in C. elegans RSKS-1 is invariably conserved in mammalian S6K (Fig. 1D); therefore, it will be interesting to determine whether the orthologous E to K change in S6K extends lifespan in mice or is associated with human longevity.
Methods
Request a detailed protocolAmino acid sequence alignment of S6 kinase proteins in human, mouse, fly, and worm was conducted by using the ClustalX2 program (Thompson et al. 1997). The structural model of C. elegans RSKS-1 was generated with ModBase (Pieper et al. 2014) and subsequently visualized by using PyMOL program (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC., https://pymol.org/2/).
Lifespan assays were performed as previously described (Park et al., 2020). All lifespan measurements were performed from day 1 of adulthood. Animals were cultured at 20°C on nematode growth medium (NGM) agar plates seeded with Escherichia coli strain OP50. One hundred μl of OP50, which was cultured overnight at 37°C in liquid Luria broth (LB) media with 10 μg/ml streptomycin, was seeded on 35 mm NGM agar plates. Worms were synchronized at 20°C from hatching, and young adult worms were transferred to fresh OP50-seeded NGM plates. The worms were transferred again to fresh OP50-seeded plates every day until the worms stopped laying embryos. The deaths of worms were scored every two days. Worms that did not respond to a gentle touch with a sterilized platinum wire were determined as dead. Animals that burrowed, displayed internal hatching or vulval rupture, or crawled off the plates were censored but included in the statistical analysis.
Lifespan curves were generated and the statistical analysis was performed by using OASIS2 (https://sbi.postech.ac.kr/oasis2/) (Han et al., 2016). p-values were calculated using a log-rank (Mantel-Cox method) test. The lifespan curves were pooled from two or three independent lifespan data obtained by two researchers.
Reagents
C. elegans strains used in this study are as follows: wild-type Bristol N2, IJ1970 rsks-1(gk185793), IJ2046 rsks-1(gk587386), IJ109 rsks-1(tm1714), and IJ1954 rsks-1(gk902595). All mutant strains were outcrossed four times to Lee laboratory N2.
Acknowledgments
We thank the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure (P40 OD010440), for providing strains. We also thank all the Lee lab members for comments on the manuscript and discussion.
References
Funding
This study is supported by the Korean Government (MSICT) through the National Research Foundation of Korea (NRF) (NRF-2019R1A3B2067745) to S‐J.V.L.
Reviewed By
Meng-Qiu Dong and AnonymousHistory
Received: September 3, 2020Revision received: October 19, 2020
Accepted: October 26, 2020
Published: October 27, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Lee, J; Kim, SS; Lee, SJV (2020). A mutation that alters the 255th glutamate to lysine in RSKS-1/S6 kinase reliably extends the lifespan of C. elegans. microPublication Biology. 10.17912/micropub.biology.000322.Download: RIS BibTeX