Department of Genetics, Yale School of Medicine, New Haven, CT 06519, USA
Yale College, New Haven, CT 06511, USA
Department of Cell Biology, Yale School of Medicine, New Haven, CT 06519, USA
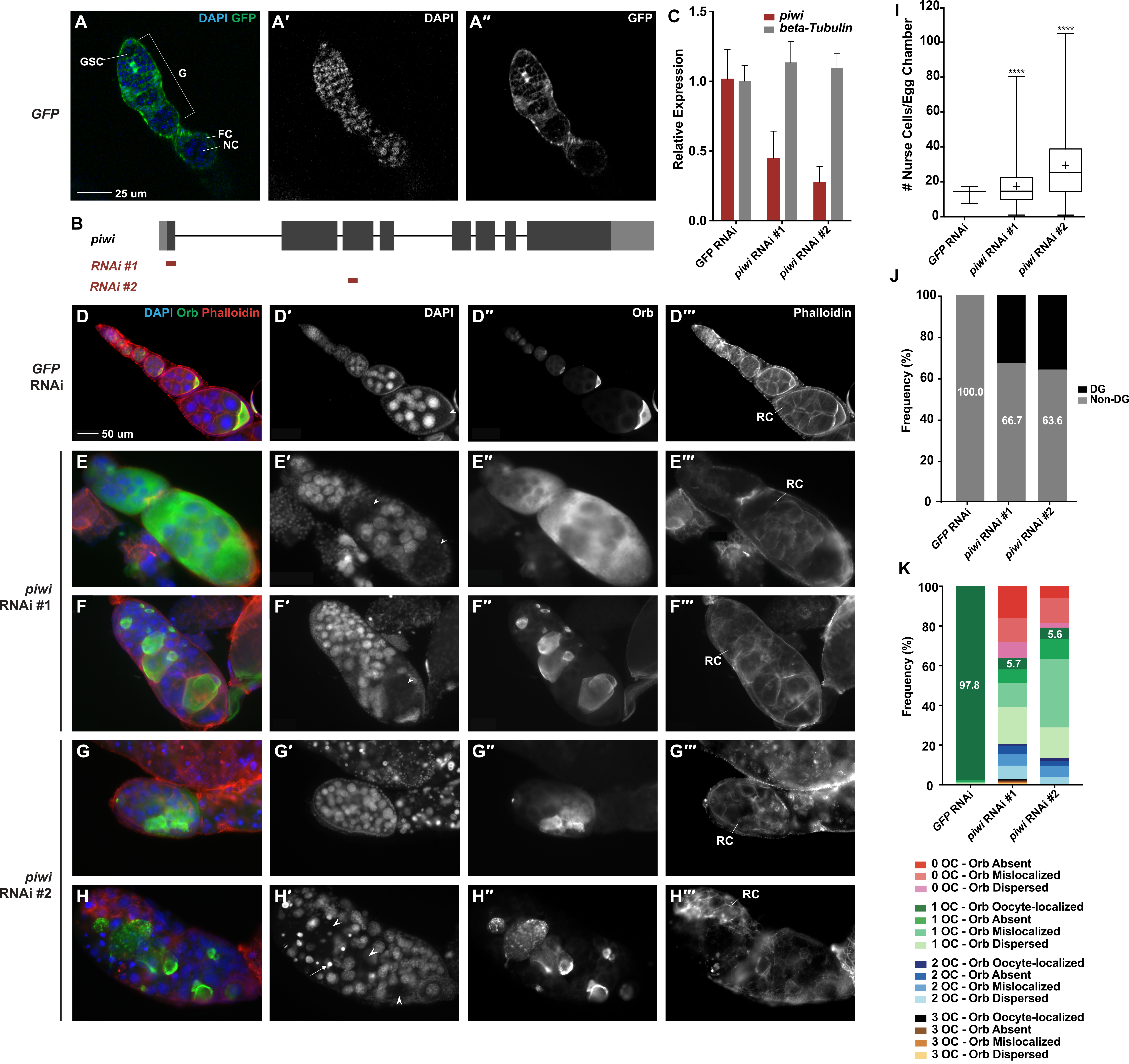
(A – A′′) tj-GAL4 driving UASp-GFP demonstrates that the driver is expressed in all ovarian somatic cells. (B) A schematic drawing of piwi gene structure and target regions of piwi RNAi #1 and piwi RNAi #2. (C) RT-qPCR demonstrates that tj-GAL4 driving piwi RNAi #1 or piwi RNAi #2 reduces piwi expression in all ovarian somatic cells compared to the control. (D – H′′′) tj-GAL4 RNAi experiments show that piwi depletion in ovarian somatic cells causes dramatic morphological defects. DAPI staining marks the nuclei of nurse cells and presumptive oocyte nuclei, Orb is the oocyte-specific protein, and Phalloidin marks F-actin structures, including ring canals. Presumptive oocytes are indicated with arrowheads. Degenerate nurse cells, identified by pyknotic nuclei, are indicated with arrows. (D – D′′′) tj-GAL4 driving GFP RNAi demonstrates the normal egg chamber morphology of 15 nurse cells, 1 presumptive oocyte, and oocyte-localized Orb staining. (E – E′′′) 15 nurse cells are observed, but two presumptive oocytes are identified. The Orb protein localization pattern is dispersed throughout the egg chamber. (F – F′′′) A tumorous number of nurse cells are observed and one presumptive oocyte is observed. However, the Orb protein is mislocalized to non-oocyte germline cells. (G – G′′′) A tumorous number of nurse cells is observed and no presumptive oocytes are observed; however, Orb staining is still observed and is mislocalized. (H – H′′′) Degenerate nurse cells are observed, identifiable by their bright pyknotic nuclei. No presumptive oocyte is observed, but Orb staining is still present and thus considered mislocalized. (I) A box and whisker plot of the number of nurse cells per egg chamber for each genotype. **** represents a p-value < 0.0001 compared to tj-GAL4>GFP RNAi, unpaired t-test. (J) The frequency of egg chambers that had non-degenerate and degenerate nurse cells. (K) The frequency of egg chambers with zero, one, two, or three presumptive oocytes (OC) and with oocyte-localized, absent, mislocalized, or dispersed Orb staining. The number of egg chambers counted for each genotype is as follows: tj-GAL4>GFP RNAi = 1210; tj-GAL4>piwi RNAi #1 = 159; tj-GAL4> piwi RNAi #2 = 213. DG=degenerate, F= follicle cell, G=germarium, GSC=germline stem cell, NC=nurse cell, OC=oocyte, RC=ring canal.
Description
The development of germ cells requires communication between the germline and the soma, a process that has been extensively characterized within the Drosophila ovary. Throughout the lifetime of the female fly, germline stem cells (GSCs) continually self-renew and produce differentiating daughter cells (cystoblasts), and this process relies on communication with the ovarian somatic cells (OSCs) within the stem cell niche: terminal filament cells, cap cells, and escort cells (Lin and Spradling 1993, Xie and Spradling 2000). Cystoblasts then go through four rounds of cell division with incomplete cytokinesis to produce a germline cyst of precisely 16 sister cells (cystocytes) with their cytoplasm connected by intercellular cytoplasmic bridges called ring canals. One of the 16 cystocytes subsequently becomes the oocyte, while the other 15 become nurse cells. Nurse cells endoreplicate to become extremely polyploid, and produce and transport mRNAs and proteins into the oocyte via ring canals to promote its maturation. Due to this unidirectional flow, the oocyte can be easily identified even within the earliest stage egg chambers by the localization of proteins such as Orb (Christerson and McKearin 1994, Lantz et al. 1994), and at later stages it is easily recognizable from its very small nucleus-to-cytoplasm ratio. This 16-cell germline cyst is encapsulated by a monolayer of somatic follicle cells, which support oocyte development throughout the remainder of oogenesis.
Early in the characterization of the stem cell niche, genetic screens revealed that Piwi, the founding member of the evolutionarily conserved PIWI/AGO protein family which is now well-known for its transposon suppression function, is required for the maintenance of GSC stem-ness (Cox et al. 1998, Cox et al. 2000). The ovaries of null piwi mutants lack germline cells almost entirely, reflecting a role for piwi in GSC self-renewal. Mosaic analysis showed that piwi mutations in germline cells of the ovary did not impair GSC self-renewal (Cox et al. 1998), suggesting that piwi expression in the ovarian soma is required for GSC self-renewal, and thereby implicating Piwi in soma-to-germline crosstalk.
The vast majority of further studies into the functions of piwi and its associated small noncoding PIWI-interacting RNA (piRNA) have focused on germline cells. It has been discovered that Piwi largely localizes to the nucleus, where it is guided by piRNA to target nascent RNA. There, Piwi impacts gene expression by modulating chromatin modifications (Brower-Toland et al. 2007, Wang and Elgin 2011, Sienskiet et al. 2012). Piwi and other PIWI proteins can also regulate gene expression post-transcriptionally to influence a variety of developmental processes (Robine et al. 2009, Saito et al. 2009). In addition to the many studies into the role of Piwi within the Drosophila germline, some studies have investigated its ovarian somatic function. Two such studies revealed that piwi expression in escort cells prevents the formation of GSC tumors (Jin et al. 2013, Ma et al. 2014), indicating that somatic Piwi can influence the differentiation of germline cells.
To further investigate the function of Piwi in OSCs in regulating germline cells, we used traffic jam GAL4 (tj-GAL4), which expresses in all somatic cells but not germline cells of the ovary (Fig 1 A- A′′, Olivieri et al. 2010), to deplete piwi expression in all OSCs (herein abbreviated as piwi-sKD). The two anti-piwi RNAi lines we used, “piwi RNAi #1” and “piwi RNAi #2,” target different regions of the piwi mRNA (Figure 1B) and result in different degrees of piwi knockdown (Figure 1C), providing an opportunity to explore Piwi’s ovarian somatic function in an expression-level-dependent way. We will refer to the piwi somatic knockdown as piwi-sKD #1 and piwi-sKD #2.
Piwi-sKD resulted in a variety of defects, including an abnormal number of nurse cells, an increase in the number of degenerate nurse cells, failure of proper oocyte specification, failure of proper Orb localization, loss of polarity in the egg chamber, and a general disorganization of ovarioles (cf. Fig. 1D and 1E – H′′′). We observed these defects in very early as well as late stage egg chambers, but limited the quantification of these defects to stage 6-10 egg chambers, because this is when the oocyte becomes morphologically recognizable (a “presumptive oocyte”).
Defects in nurse cell number ranged dramatically: in piwi-sKD ovaries, we observed egg chambers with as few as one nurse cell and others with over 100 nurse cells, with the average number significantly above the typical 15 nurse cells in a wild-type egg chamber (Fig 1I). Strikingly, over 30% of piwi-sKD egg chambers were “tumorous,” containing more than 30 nurse cells, some of which appeared to be diploid (Fig 1I). We also observed an increased frequency of egg chambers containing degenerate (DG) nurse cells in piwi-sKD egg chambers compared to controls (Fig 1J). These nurse cell number abnormalities suggest a role for somatic Piwi in regulating germline cystoblast and/or cystocyte proliferation.
In addition, many piwi-sKD egg chambers were abnormal in number and/or specification of oocytes. The number of presumptive oocytes per egg chamber ranged from zero to three. Among these presumptive oocytes, some had correct Orb protein localization (“oocyte-localized”), but other egg chambers lacked any Orb staining (“absent”), contained Orb protein localized to non-oocyte cells (“mislocalized”), or contained Orb protein dispersed throughout the whole egg chamber (“dispersed”). Only 5.7% and 5.6% of piwi-sKD #1 and piwi-sKD #2 egg chambers, respectively, had one oocyte and oocyte-localized Orb (Fig 1K), indicating that somatic Piwi plays a role in oocyte specification.
The defects in nurse cell number and oocyte specification upon somatic piwi depletion reveal novel roles for Piwi in OSCs in regulating the proliferation of germline cells and the specification of the oocyte. This function is distinct from the well-described role of somatic Piwi in GSC maintenance, wherein the loss of Piwi function results in a loss of germline cells altogether. Instead, this piwi-sKD phenotype echoes the germline over-proliferation phenotypes observed upon depleting piwi expression in testicular somatic cells (Gonzalez et al. 2015) or ovarian escort cells (Jin et al. 2013, Ma et al. 2014).
It is possible that some of the defects we have observed upon piwi-sKD are the result of GSC tumors observed by other groups upon piwi depletion in escort cells. Typically, a GSC, marked by a cytoskeletal structure called the spectrosome, produces 16-cell cysts connected via the fusome, a branched inter-cellular derivative structure of the spectrosome that traverses ring canals within an egg chamber. The GSC tumor-filled germaria formed by piwi depletion in escort cells were described as containing mostly undifferentiated germ cells marked with spectrosomes (Jin et al. 2013, Ma et al. 2014). The tumorous egg chambers we observed upon piwi-sKD may result from the encapsulation of these GSC tumors by follicle cells. However, in the majority of egg chambers, we observe polyploid germ cells that are connected by ring canals, so some cystoblasts must have undergone endoreplication and cell division with incomplete cytokinesis. The fact that the piwi-sKD egg chambers often contain many more than 15 nurse cells suggests over-proliferation of cystocytes within a cyst or incorporation of multiple cysts into the same egg chamber, or both.
The possibility that at least some of the aberrant piwi-sKD egg chambers contain multiple germline cysts is supported by our observation of many egg chambers with multiple oocytes. However, this explanation likely does not extend to egg chambers containing no presumptive oocyte or dislocalized Orb in piwi-sKD egg chambers. In those egg chambers, oocytes are not correctly positioned within the egg chamber, and either oocytes are not properly specified by signaling from follicle cells, or the transportation of oocyte-specific markers, like Orb, into the oocyte does not occur.
Overall, our study reveals the function of piwi in somatic cells of the ovary to regulate germline cyst formation and oocyte specification. More cell-type specific piwi depletion will further reveal whether Piwi functions in particular sub-types of ovarian somatic cells to regulate the above soma-to-germline signaling during oogenesis.
Methods
Request a detailed protocolDrosophila husbandry and genetics
All Drosophila stocks were raised on standard agar/molasses medium and raised at 25°C for experiments. Traffic jam-GAL4 (tj-GAL4) from Kyoto Stock Center (DGRC #104055) was used to express UASp constructs in all somatic cells of the ovary. UASp-GFP (BDSC #1521) was used to verify the expression pattern of tj-GAL4. Two anti-Piwi RNAi lines (“piwi RNAi #1” and “piwi RNAi #2” in this study) were used to knock down piwi expression. piwi RNAi #1 targets exon 2 of the piwi mRNA and is BDSC stock #37483; piwi RNAi #2 targets exon 3 of the piwi mRNA and was a gift from T. Xie, Stowers Institute For Medical Research, Kansas City, MO. GFP RNAi (BDSC stock #41550) was used as a negative control. To generate GAL4/UASp flies for analysis, two males carrying the GAL4 driver were crossed with three virgin females carrying the UASp construct. GAL4/UASp females were aged for two to three days in a ratio of 2:1 with w1118 males prior to ovary dissection.
RNA extraction and RT-qPCR
Ovaries were dissected from 2-3 day-old females in 1XPBS, then transferred to TRIzol (Thermo Fischer Scientific, #15596018) and RNA was extracted following manufacturer’s instructions. 6-8 pairs of ovaries were grouped together for each sample. 2 µg of total RNA was used for reverse transcription using MultiScribe (Thermo Fischer Scientific, #4311235) following manufacturer’s instructions, and diluted 5x before quantitative PCR using iTaq SYBR MasterMix (BioRad, #172-5125). Relative fold changes were calculated using the ΔΔCT method, with actin5C as the reference (F: GAAGAAGTTGCTGCTCTGGTTG; R: GAGCATCGTCTCCGGCAAATC), to determine expression levels of piwi (F: TTGTCCAATCATGCTAACCTTCTGGGAT, R: CTTATTTCGATCTTAGCTCGGGGATC) and beta-Tubulin (F: TGTCGCGTGTGAAACACTTC, R: AGCAGGCGTTTCCAATCTG).
Immunostaining
Ovaries from 2-3 day old females were dissected in 1X PBS and fixed with 200 μL of the following fixing solution (v/v%): PBS (89.5%), 10% Nonidet P-40 (5%), 37% formaldehyde (5.5%). The fixed ovaries were then washed 3 times for 15 minutes each in PBST (PBS and 0.2% Triton X-100). Ovaries were then blocked overnight in 5% NGS at 4oC, followed by incubation overnight at 4oC in primary antibody diluted in PBST + 2% NGS. Samples were washed three times in PBST and incubated in secondary antibodies overnight at 4oC. Samples were washed three times with PBST, stained with DAPI (1:500) and Phalloidin (1:200, ThermoFisher, #R415) for 15 minutes, and mounted in Vectashield mounting media (Vector Labs, #H1000).
The following primary antibodies were used: mouse anti-Piwi 4K5 (1:15,000, made in Lin Lab) and mouse anti-Orb 4H8 (1:300, DSHB). The following conjugated secondary antibodies were used, all at 1:500 dilutions: The Alexa 488-conjugated goat anti-mouse antibody and the Alexa 555-conjugated goat anti-mouse. Orb, Phalloidin, and DAPI were used to qualitatively and quantitatively characterize the observed phenotypes of the egg chambers from all crosses.
Microscopy and phenotypic characterization
To observe the GAL4 expression pattern, confocal images of DAPI stained tj-GAL4>UASp-GFP ovaries were taken using Leica TCS SP5 Confocal Laser Scanning Microscope. Piwi knockdown samples were analyzed on the ZEISS Axio Imager2 for quantitative and qualitative characterizations.
DAPI and Phalloidin staining was used to identify nurse cells and oocytes. To more accurately quantify the effect of piwi-sKD on the proliferation and development of nurse cells, we created four categories to describe how many nurse cells were in an egg chamber: 15 normal nurse cells; 0-14 normal nurse cells; 16-29 normal nurse cells; and ≥30 normal nurse cells, which we referred to as a tumorous number. Degenerate nurse cells were identified by the presence of bright pycnotic nuclei. Any large area of cytoplasm without a polyploid nucleus was considered a presumptive oocyte. We quantified how many egg chambers had zero, one, two, or three presumptive oocytes. We also characterized the localization of the Orb staining based on its relative position with the presumptive oocyte. We created four categories of Orb’s localization pattern: absent, oocyte-localized, mislocalized, and dispersed. If the egg chamber had no trace of Orb staining, the pattern was considered absent. If the Orb staining localized with the presumptive oocyte(s), it was designated oocyte-localized; but if the Orb staining was not localized to the presumptive oocyte and formed discrete patches, it was considered mislocalized. If the Orb staining filled the entire egg chamber, its pattern was considered dispersed.
Statistical Analysis
Statistical significance for nurse cell number in piwi-sKD was assessed via an unpaired t-test using tj-GAL4 driving GFP siRNA as a control.
Acknowledgments
We acknowledge the Developmental Studies Hybridoma Bank for providing antibodies, the Bloomington Drosophila Stock Center, Kyoto Stock Center, and Ting Xie for providing fly stocks, and members of the H. Lin Lab for discussion and technical support.
References
Funding
This work was funded by a gift from Luye Life Sciences to Haifan Lin. Gina Zhu was supported by The Yale College Dean’s Research in the Sciences Fellowship, The John E. Linck and Alanne Headland Linck Fellowship, and the Gary Stein Fellowship. Lauren E Gonzalez was supported by NSF Graduate Research Fellowship Program (DGE1752134) and the Training Program in Genetics (NIH T32 GM007499).
Reviewed By
AnonymousHistory
Received: August 23, 2020Revision received: October 23, 2020
Accepted: October 28, 2020
Published: November 18, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Gonzalez, LE; Zhu, G; Lin, H (2020). Ovarian somatic Piwi regulates nurse cell proliferation and oocyte specification in Drosophila. microPublication Biology. 10.17912/micropub.biology.000324.Download: RIS BibTeX