Abstract
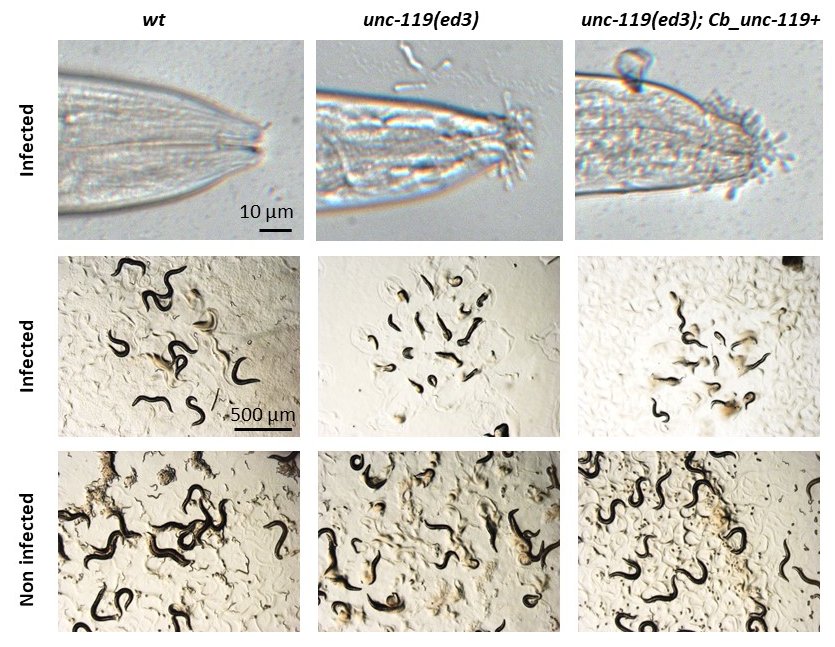
If the cuticle acts as a protective barrier against environmental insults, several pathogens have developed strategies that use it as a way to infect C. elegans. The fungus Drechmeria coniospora produces spores that attach to the cuticle, before hyphae invade the body. Mutants with an altered surface coat, the outermost layer of the cuticle, including bus-2, bus-4, bus-12 and bus-17 show increased adhesion of fungal spores (Rouger et al, 2014; Zugasti et al, 2016). We unexpectedly found that D. coniospora spores attach unusually densely around the mouth of unc-119 mutants. Interestingly, this phenotype is not rescued by the C. briggsae unc-119 construct that is conventionally used to rescue neuronal unc-119 phenotypes.
Description
As part of our investigations of the interaction between C. elegans and D. coniospora, we made use of MosSCI strains, constructed in an unc-119(ed3) background (Frøkjær-Jensen et al., 2008; Maduro, 2015). We noticed that a variety of these strains exhibited a greatly increased susceptibility to infection. Upon further examination, we determined that this was due to an increase in the adhesion of fungal spores, most prominently at the tip of the head (Figure 1). The phenotype was not observed in a strain carrying a wild-type C. elegans unc-119 rescuing construct in an unc-119(e2498) background. But the increased spore adhesion was visible in both the unc-119(ed3) and the unc-119(tm4063) background even in the presence of the standard C. briggsae unc-119 rescuing construct. The phenotype was absent from these same transgenic strains in which unc-119(ed3) was eliminated by out-crossing (Table 1 below). unc-119 function has been extensively analysed in the nervous system. Notably, some expression in the epidermis was reported recently (Lear et al., 2018). While we have not determined the precise cause, since spore adhesion is a major determinant of infection progression (Zugasti et al., 2016), such effects need to be taken into account when interpreting experiments involving any strain that has an unc-119 allele in it, which has been often employed as selectable marker for transgenesis.
| strain | genotype | spore adhesion |
| N2 | wt | normal |
| EG6699 | unc-119(ed3) III; ttTi5605 II | increased |
| IG1604 | unc-119(ed3) III; frSi6[col-154p::CEBP-1::GFP::3’cebp-1, Cb-unc-119(+) ttTi5605] II | increased |
| IG1633 | +; frSi6[col-154p::CEBP-1::GFP::3’cebp-1, Cb-unc-119(+) ttTi5605] II | normal |
| IG1622 | unc-119(ed3) III; frSi9[pNP151(col-62p::Lifeact::mKate_3’c-nmy), Cb-unc-119(+) ttTi5605] II | increased |
| IG1623 | +; frSi9[pNP151(col-62p::Lifeact::mKate_3’c-nmy), Cb-unc-119(+) ttTi5605] II | normal |
| unc-119(ed3) III; frSi10[pNP150(F40H7.12p::GFP), Cb-unc-119(+) ttTi5605] II | increased | |
| AX6672 | unc-119(ed3) III; npr-1(ad609); ilcr-1(tm5866); [ilcr-1p::loxp::ILCR-1::let-858 3’UTR , Cb-unc-119(+) ttTi5605] II | increased |
| OP533 | unc-119(tm4063) III; wgIs533[CEH-18::TY1::EGFP::3xFLAG, Cb-unc-119(+)] | increased |
| JR667 | unc-119(e2498::Tc1) III; wIs51[SCMp::GFP, Ce-unc-119(+)] V | normal |
Methods
Request a detailed protocolEggs, prepared by the standard bleach method, were allowed to hatch in 50 mM NaCl in the absence of food at 25°C overnight. Synchronized L1 larvae were transferred to NGM agar plates spread with E. coli OP50 and cultured at 25°C until the L4 stage (40 h) before being exposed to fungal spores as previously described (Pujol et al., 2001). Images were taken of worms mounted on a 2% agarose pad on a glass slide anesthetized with 0.25 mM levamisole, using a Zeiss AxioCam HR digital colour camera and AxioVision Rel. 4.6 software (Carl Zeiss AG).
Reagents
N2, EG6699 unc-119(ed3); ttTi5605 II, JR667 unc-119(e2498::Tc1) III; wIs51[SCMp::GFP, Ce-unc-119(+)] V and OP533 unc-119(tm4063) III; wgIs533[CEH-18::TY1::EGFP::3xFLAG, Cb-unc-119(+)] strains were provided by the CGC (Caenorhabditis Genetics Center), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). In addition, the following strains were tested for spore adhesion: IG1604 unc-119(ed3); frSi6[col-154p::CEBP-1::GFP::3’cebp-1, Cb-unc-119(+) ttTi5605] II (Kim et al., 2016), IG1622 unc-119(ed3); frSi9[pNP151(col-62p::Lifeact::mKate_3’c-nmy), Cb-unc-119(+) ttTi5605] II, IG1623 frSi9[pNP151(col-62p::Lifeact::mKate_3’c-nmy), Cb-unc-119(+) ttTi5605] II (Taffoni et al., 2020), AX6672 unc-119(ed3); npr-1(ad609); ilcr-1(tm5866); [ilcr-1p::loxp::ILCR-1::let-858 3’UTR, Cb-unc-119(+) ttTi5605] II (Chen et al., 2017), IG1633 frSi6[pNP145(col-154p::CEBP-1::GFP::3’cebp-1), Cb-unc-119(+) ttTi5605] II and IG1629 unc-119(ed3); frSi10[pNP150(F40H7.12p::GFP), Cb-unc-119(+) ttTi5605] II, this study. Both pNP145 and pNP150 were derived from pCFJ151 – ttTi5605_MCS, that was a gift from Erik Jorgensen (Addgene plasmid # 19330 ; http://n2t.net/addgene:19330 ; RRID:Addgene_19330) (Frøkjær-Jensen et al., 2008).
Acknowledgments
We thank Jonathan Ewbank for his detailed and constructive comments.
References
Funding
French National Research Agency (ANR-16-CE15-0001-01, ANR-16-CONV-0001) and institutional grants from CNRS, INSERM and Aix Marseille University to the CIML.
Reviewed By
AnonymousHistory
Received: December 14, 2020Revision received: December 29, 2020
Accepted: December 30, 2020
Published: January 5, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Omi, S; Pujol, N (2021). unc-119 mutants have an increased fungal spore adhesion that is not rescued by Cb-unc-119. microPublication Biology. 10.17912/micropub.biology.000344.Download: RIS BibTeX