Description
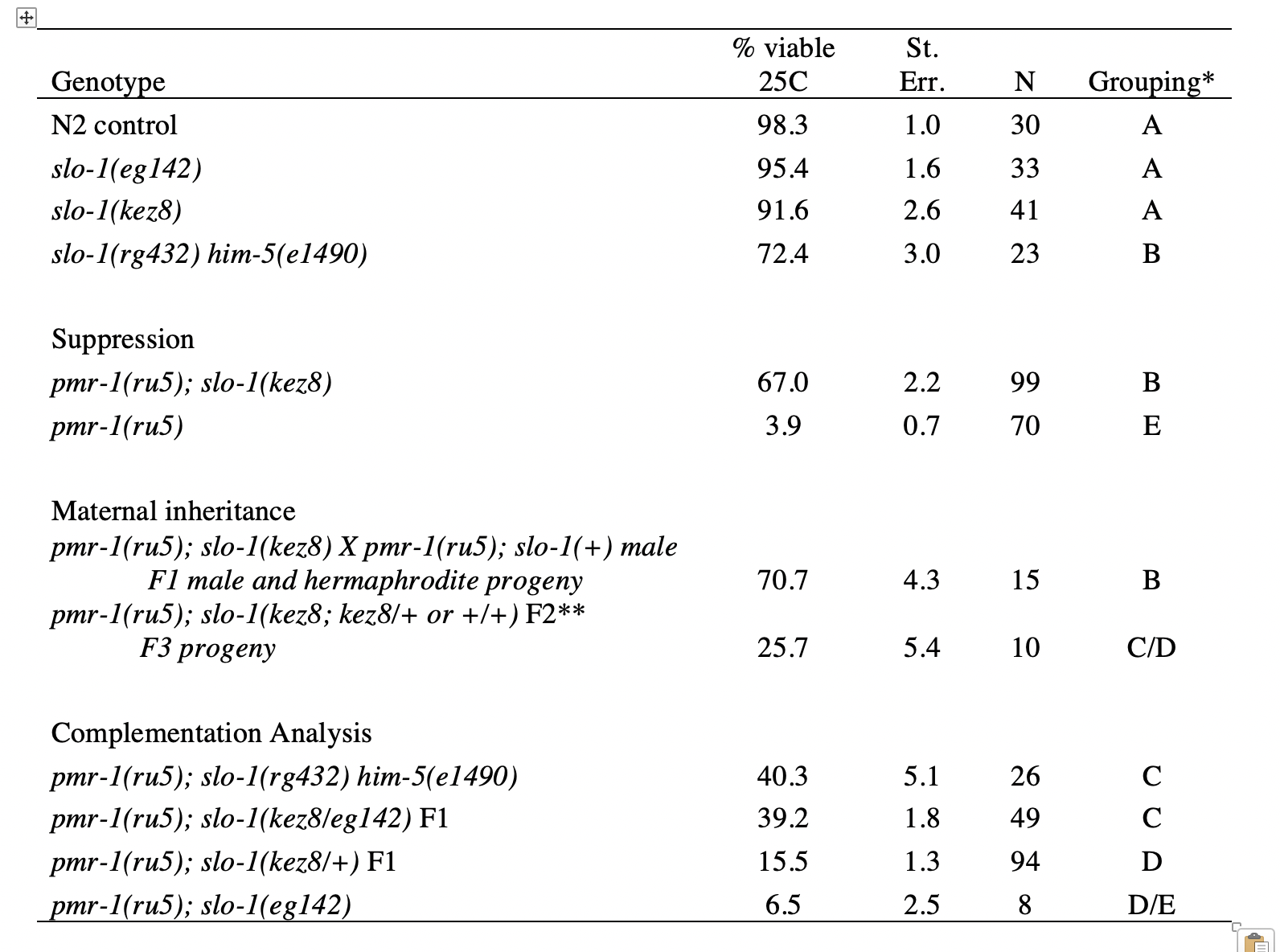
Calcium signaling is known to play a critical role in cell migration (Ridley et al., 2003; Wei et al.., 2012). In C. elegans, embryos with disruptions in the secretory pathway calcium ATPase gene pmr-1 show defects in cell migration that result in lethal phenotypes. Previous work has shown that pmr-1(lof) phenotypes can be suppressed by changes in the activity of the calcium channels IP3-receptor/ITR-1 and ryanodine receptor/UNC-68, indicating that cell migration defects are the results of changes in calcium homeostasis (Praitis et al., 2013). To identify additional genes important for cell migration during embryogenesis, we performed a genetic screen to isolate additional suppressors of the pmr-1(ru5) strain, reasoning that disruption of cell migration due to changes in pmr-1 activity could be suppressed by commensurate changes in other genes that regulate calcium levels or signaling. In this screen, we identified the kez8 allele. We confirmed that the kez8 allele suppresses the pmr-1(ru5) mutant phenotype by examining the percentage of viable progeny produced by pmr-1(ru5); kez8 strains when grown under restrictive conditions. The viability of the pmr-1(ru5); kez8 strain is 67%, significantly higher than the 3.9% viability observed in the pmr-1(ru5) control strain at 25C (Table 1; p<0.01).
Genetic inheritance analysis shows that maternal and not zygotic genotype is crucial for kez8 suppression of embryonic lethality in pmr-1(ru5) strains. pmr-1(ru5) hermaphrodites homozygous for the kez8 suppressor produced viable progeny at similar rates whether they were allowed to self-fertilize or were crossed to pmr-1(ru5) males that did not carry a suppressor allele (Table 1; p<0.01). F1 pmr-1(ru5); kez8/+ hermaphrodites from this cross produced some viable F2 progeny at rates not dissimilar to what would be expected if the allele was acting zygotically (Table 1). If zygotic genotype was critical to F2 survival, we’d predict that individuals homozygous for the kez8 allele would be over-represented in the F2 population, and these F2 hermaphrodites would produce F3 progeny with viability rates similar to the parental strain. However, when the surviving F2 progeny were allowed to reproduce, the viability of their progeny was significantly lower than the parental strain (Table 1; P<0.01). When we examined the F3 viability rates from F2 individuals, they represented each of the predicted genotypic classes (kez8, kez8/+ or +; ChiSquare, p > 0.05), indicating that zygotic genotype did not affect F2 survival. These results are consistent with a recessive maternal-effect kez8 suppressor that restores viability to pmr-1(ru5) embryos.
Mapping, sequencing, and complementation analysis shows kez8 is an allele of slo-1. We used a whole-genome sequencing (WGS) and mapping strategy to locate the suppressor to the right end of LG V, which was independently confirmed with traditional mapping. Our best candidate for the causative mutation was a single base change, from A to T, in an intron of slo-1 which maps to position 18.5 Mbp, within ~0.5 Mbp of the highest likelihood map position based on WGS analysis. The mutation was confirmed independently in pmr-1(ru5); kez8 strains by PCR amplification and sequencing and is identical to a known slo-1 allele rg432. Complementation analysis with the slo-1(eg142) nonsense allele indicates kez8 is allelic, as pmr-1(ru5); kez8/eg142 hermaphrodites produce viable progeny at levels that are significantly higher than pmr-1(ru5); kez8/+ controls (Table 1; p<0.01). However, pmr-1(ru5) strains homozygous for eg142 are not suppressed (Table 1; compare pmr-1(ru5); slo-1(eg142) to pmr-1(ru5); p=1.0) indicating that the unique nature of the kez8 allele is important for suppression. Neither the kez8 nor eg142 alleles show significant embryonic lethality in the wild type background (Table 1). To further support our identification of kez8 as an allele of slo-1, we crossed pmr-1(ru5) to a strain homozygous for slo-1(rg432), which has the same genetic change in slo-1 as kez8 but also carries a linked him-5(e1490) allele necessary for the cross but with some associated embryonic lethality in the wild type background (Hodgkin et al., 1979; Table 1). pmr-1(ru5); slo-1(rg432) him-5(e1490) hermaphrodites show significant suppression of the pmr-1(ru5) embryonic lethal phenotype (Table 1; p<0.01), consistent with our interpretation that kez8 is an allele of slo-1.
The slo-1(rg432) allele was identified as a suppressor of a male spicule protraction constitutive phenotype (PrC) caused by disruptions in the unc-103 and egl-2 EAG-like potassium channel genes. The slo-1(rg432) allele has a significant impact on mRNA expression levels for slo-1 splice variants in both males and hermaphrodites and reduces the expression levels of other genes that regulate cell excitability in males, including unc-103 and egl-2 (LeBoeuf & Garcia, 2012). Our analysis of the pmr-1(ru5); kez8 strain indicates that the kez8 allele also negatively impacts male mating. Using him-14(dsRNA), which generates a small number of male progeny that can be subsequently crossed to maintain males in a given strain (Fay, 2006), we obtained pmr-1(ru5); kez8 males. However, unlike in the parental pmr-1(ru5) strain or in other strains undergoing him-14(dsRNA) treatment, we were unable to establish stable male lines in the pmr-1(ru5); kez8 strain, indicating that the kez8 allele is associated with low male mating efficiency.
SLO-1 is a BK-type calcium-sensitive potassium channel with established roles in C. elegans in regulating neurotransmitter release, ethanol intoxication and male mating behavior (Salkoff et al.., 2005; LeBoeuf & Garcia, 2012). Our results reveal a new role for slo-1 in C. elegans embryonic cell migration. Given that slo-1(kez8), as well as alleles of other genes identified as genetic interactors of pmr-1 including IP3-receptor/itr-1 and ryanodine receptor/unc-68, also impact male mating behavior (Garcia et al. 2003; Gower et al. 2005; LeBoeuf & Garcia, 2012; Praitis et al. 2013), a set of similar genes may be involved in both processes. A simple model for the interaction between PMR-1 and SLO-1 is that elevated cytosolic calcium predicted in pmr-1(lof) mutants disrupts the normal regulation of BK channel activity during migration, which is suppressed in the kez8/rg432 mutants because of changes in overall or isoform-specific slo-1 expression levels (LeBoeuf & Garcia, 2012). The finding that changes in slo-1 activity impact embryonic cell migration in C. elegans is consistent with previously identified roles for SLO/BK-type channels in the migration and invasive properties of glioblastomas (Catacuzzeno et al., 2015; Ge et al.., 2014).
Methods
Request a detailed protocolWe performed a forward genetic screen to identify recessive suppressors of the pmr-1(ru5) strain (Praitis et al.., 2013) using a MosI transposon mutagenesis protocol (Boulin & Bessereau, 2007). As we could not identify the MosI insertion site, suppressors were mapped using a modified whole genome sequencing (WGS) mapping strategy (Minevich et al., 2012) where we backcrossed suppressor lines to CB4856 and then used polymorphisms in the Illumina sequenced DNA to locate and identify the causative mutation. The mapped location and sequence change were independently verified using conventional mapping strategies and PCR amplification followed by sequencing. We determined the number of viable progeny produced by single hermaphrodites for all strains, as previously described (Praitis et al.., 2013). We used conventional approaches for complementation and inheritance analysis (Brenner, 1974; Fay, 2006; Fay, 2013).
Request a detailed protocol.
Reagents
We used the following strains: N2, AZ5 pmr-1(ru5) I, CG925 slo1(rg432) him-5(e1490) V, OH441 otIs45 V, SK4005 zdIs45 I, BZ142 slo-1(eg142) V, CB2065 dpy-11(e224) unc-76(e911) V, GRN83 pmr-1(ru5) I; kez8, GRN171 pmr-1(ru5) zdIs5 I, GRN414 pmr-1(ru5); zdIs45 I; slo-1(eg142) V. Lab generated strains were backcrossed to N2 4 to 6 times. For whole genome sequencing and mapping analysis, lab-generated strains were backcrossed 4 to 6 times to strains with a CB4856 background: GRN229 pmr-1(ru5) zdIs5 I, GRN230 pmr-1(ru5) zdIs5 I, GRN231 pmr-1(ru5) zdIs5 I, GRN232 pmr-1(ru5) zdIs5 I, GRN292 pmr-1(ru5) zdIs5 I; kez8, GRN293 pmr-1(ru5) zdIs5 I; kez8, GRN336 pmr-1(ru5) zdIs5 I; kez8. The strains GRN395 kez8, GRN406 kez8, and GRN407 kez8 were derived from parents with CB4856 and N2 backgrounds. All of the GRN strains are from the Praitis lab, as is AZ5. The CG925 strain is from the Garcia lab. Other strains were obtained from the CGC.
Acknowledgments
We thank the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure (P40 OD010440), and the Garcia lab for providing strains.
References
Funding
We thank Grinnell College for its support of this research through the Support for Faculty Scholarship fund and the Grinnell College MAP program.
Reviewed By
AnonymousHistory
Received: November 13, 2020Revision received: December 23, 2020
Accepted: January 10, 2021
Published: January 14, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Thurman, M; Sun, H; Kubica, S; Praitis, V (2021). The slo-1 BK potassium channel interacts genetically with pmr-1 secretory pathway calcium ATPase during C. elegans embryonic cell migration. microPublication Biology. 10.17912/micropub.biology.000351.Download: RIS BibTeX