Department of Biology
Indiana University
The University of North Carolina, Chapel Hill
Abstract
We have previously adapted a select number of Drosophila cell lines to grow in serum-free media supplemented with fly extract. This condition is arguably more representative of a native growth environment. Here, we validated that the fly extract adapted line, S2R+ (FEx 2.5%) is amenable to RNAi. RNAi against Rho1 in both S2R+ and S2R+ (FEx 2.5%) produced phenotypes similar to ones previously described in Drosophila S2 cells.
Description
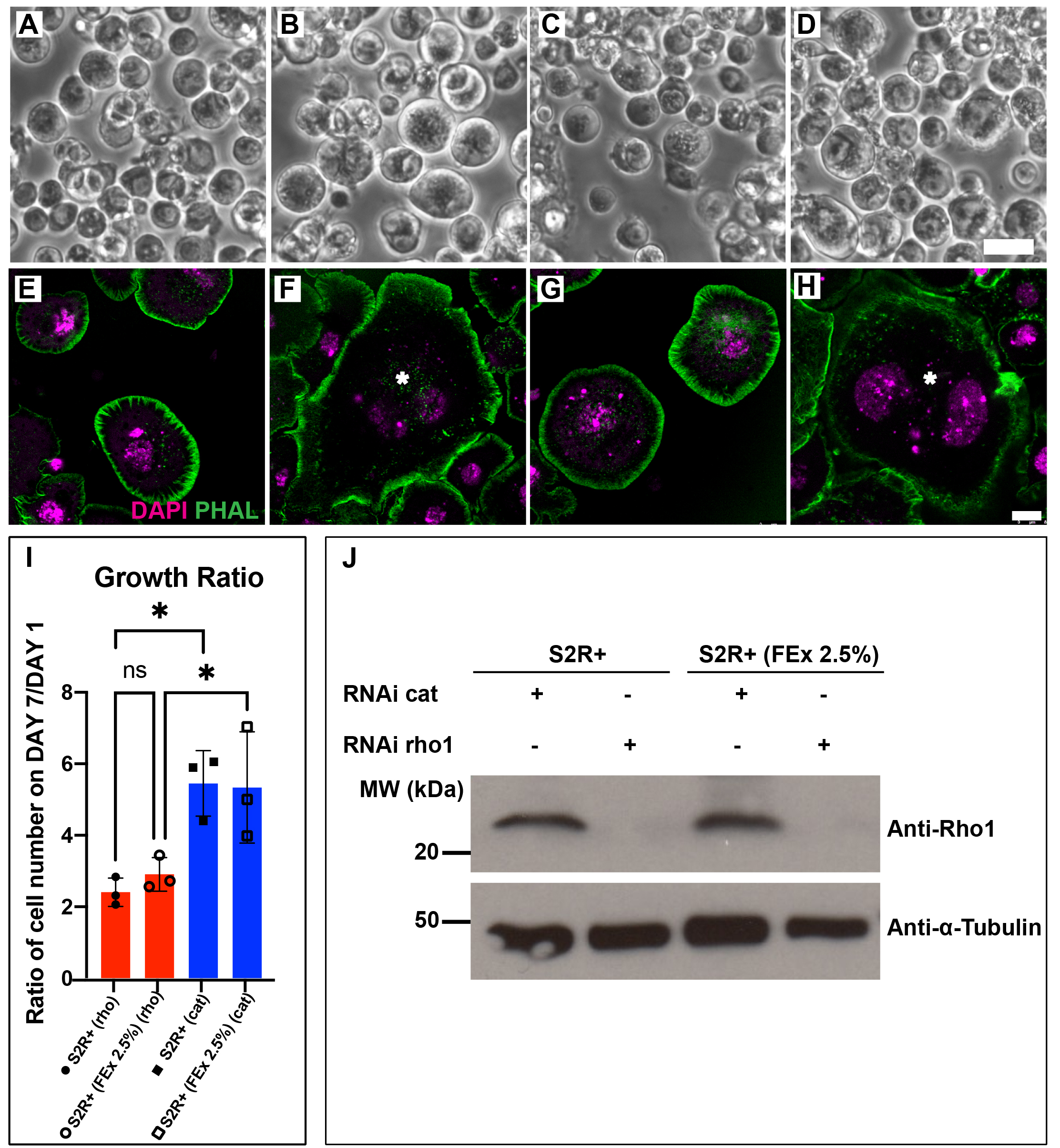
Drosophila Schneider S2 cell lines are susceptible to RNA interference (RNAi) and this attribute has cemented Drosophila cell lines as an important tool for high throughput functional genomics screening (Rogers and Rogers 2008; Zhou et al. 2013; Mohr 2014). RNAi against Drosophila Rho1 in S2 cells results in a block in mitosis, giving rise to enlarged and multinucleated cells (Rogers et al. 2004). Recently, we have adapted a select group of Drosophila embryonic cell lines to grow in media supplemented by adult fly extract (FEx), instead of fetal bovine serum (FBS) (Luhur et al. 2020). Here, we demonstrate that S2R+ (FEx 2.5%), the M3 + 2.5% FEx-adapted S2R+ line is also amenable to RNA interference (RNAi), similar to its parental S2R+ cells cultured in M3 BPYE + 10% FBS. We observed similar efficacious RNAi against Rho1 in S2R+ and S2R+ (FEx 2.5%) as the cells became enlarged (Figure 1A-D), multinucleated (Figure 1E-H) and failed to proliferate (Figure 1I). There was a comparable growth delay in S2R+ and S2R+ (FEx 2.5%) cells treated with Rho1 dsRNA, as the cell population doubled in 7 days (Figure 1I). In contrast, both S2R+ and S2R+ (FEx 2.5%) cells treated with double stranded RNA against a control target gene encoding the bacterial antibiotic resistance gene chloramphenicol acetyl transferase (cat) had significantly proliferated 5 fold more under similar conditions (Figure 1I). In addition, there were no significant differences in the growth ratio between S2R+ and S2R+ (FEx 2.5%) (Figure 1I) (Luhur et al. 2020). These results demonstrate that Rho1 RNAi was recapitulated robustly in S2R+ (FEx 2.5%), similar to its parental S2R+ cells. Lastly, to confirm the depletion of Rho1, we assayed for Rho1 protein levels in these cultures by Western blot. Our result indicated a strong reduction in the amount of Rho1 protein in both S2R+ (FEx 2.5%) and S2R+ cells after Rho1 knockdown (Figure 1J). In contrast, the control RNAi knockdown of cat did not affect Rho1 protein levels in either S2R+ or S2R+ (FEx 2.5%) (Figure 1J). In summary, this finding expands the utility of the fly extract-adapted cells for their use in functional genomics in a more physiologically relevant culture condition.
Methods
Request a detailed protocolCell culture
S2R+ (DGRC#150, FBtc0000150), S2R+ (FEx 2.5%) (DGRC#310, FBtc0000310) were cultured in M3 + BPYE + 10% FBS and M3 + 2.5% FEx, respectively, according to previously described protocol (Luhur et al. 2019).
RNA interference (RNAi)
Cells from the respective growth media were pelleted and then seeded at 1 million cells/ mL in serum free M3 media in a 24 well plate (1 mL per well). 10 mg/mL dsRNA was added slowly to the media and allowed to incubate at room temperature for 1 hour. After the incubation period, the M3 media was supplemented with equal volumes of either M3+BPYE+20% FBS or M3 + 5% FEx, to constitute the M3 + BPYE + 10% FBS and M3 + 2.5% FEx, respectively. Cells were allowed to grow for a week at 25°C before assaying for the loss of function phenotypes.
As a negative control, a 467-bp fragment of the chloramphenicol resistance cassette was amplified from pFastBacHT-CAT expression plasmid (Invitrogen) using the primers: T7-CAT-fwd: 5′-TAATACGACTCACTATAGGATCCCAATGGCATCGTAAAGAACATTTTGAGGC-3′ and T7-CAT-rev:
5′-TAATACGACTCACTATAGGGGGCGAAGAAGTTGTCCATATTGGCCA-3′.
As a positive control, we amplified a 667-bp sequence for Rho1(FBgn0014020) using the primers: T7-Rho1-fwd: 5′-TAATACGACTCACTATAGGTTTGTTTTGTGTTTAGTTCGGC-3′ and T7-Rho1-rev: 5′-TAATACGACTCACTATAGGATCAAGAACAACCAGAACATCG-3′, from a Rho1 expression construct, originally provided by Dr. Liqun Luo (Stanford University). The dsRNA synthesis protocols followed the protocol described (Rogers and Rogers 2008).
Immunostaining and microscopy
The cells were seeded on dishes coated with Concanavalin A. After one hour, the media was removed and the cells were fixed for 10 minutes in a solution containing 4% paraformaldehyde diluted in phosphate buffered saline (PBS). The cells were then rinsed in 0.1% PBS-Triton-X and incubated in 1:1000 phalloidin for two hours at room temperature. Subsequently, the cells were rinsed for three times in 0.1% PBS-Triton-X before being mounted on Vectashield mounting media containing DAPI (H-1300). Fluorescence imaging was carried out using the Leica SP8 confocal microscope.
Protein extraction and Western Blotting
Cell pellets from a single well of a 24-well plate cells subjected to RNAi against either Rho1 or cat (dsRNA control) were lysed with RIPA buffer. Proteins in the lysed samples were separated with a BioRad WGX 4-20% gel, transferred onto Nitrocellulose membrane (BioRad) and blotted with either mouse anti-Rho1 (p1D9, from Developmental Studies Hybridoma Bank deposited by Parkhurst, S) or mouse anti- α-tubulin (T9026, Sigma). The blots were treated with anti-mouse HRP and the signals were visualized using Pierce Enhanced Chemiluminescence Reagent (ThermoFisher). The experiment was conducted in duplicate.
Cell counting and statistical analysis
Live cells were counted using an automated cell counter (BIORAD) according to manufacturer’s instructions. Each condition had a total of three replicate counts. Statistical analysis of the differences in the growth ratio was carried out using Prism8 using ordinary-one way ANOVA test, with Sidak’s multiple comparison test.
Acknowledgments
The Rho1 antibody was procured from Developmental Studies Hybridoma Bank (University of Iowa, IA). We also acknowledge the Light Microscopy Imaging Center (LMIC) at Indiana University Bloomington.
References
Funding
This work was supported by a NIH grant (2P40OD010949) awarded to the Drosophila Genomics Resource Center.
Reviewed By
AnonymousHistory
Received: January 5, 2021Revision received: January 22, 2021
Accepted: January 22, 2021
Published: January 29, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Luhur, A; Mariyappa, D; Klueg, KM; Rogers, SL; Zelhof, AC (2021). Serum-free adapted Drosophila S2R+ line is amenable to RNA interference. microPublication Biology. 10.17912/micropub.biology.000362.Download: RIS BibTeX