Department of Neuroscience, Brown University, Providence, RI 02912, USA
Abstract
Although some RNA-binding proteins are known to contribute to neurodegeneration, the genetic interaction between the genes encoding these proteins is unclear. Here, we examine the interaction between sym-2, the gene encoding an ortholog of hnRNPF and hnRNPH, and hrpa-1, the ortholog of of the gene encoding hnRNPA2, which when mutated causes multisystem proteinopathy. We find that after 22 hours, but not 4 hours, of paraquat-induced oxidative stress, sym-2(mn617) has a mild glutamatergic neurodegeneration phenotype. Interestingly, this defect is rescued by expression of chimeric WT hrpa-1, but not mutant. Thus, we identify a curious genetic interaction between sym-2 and hrpa-1.
Description
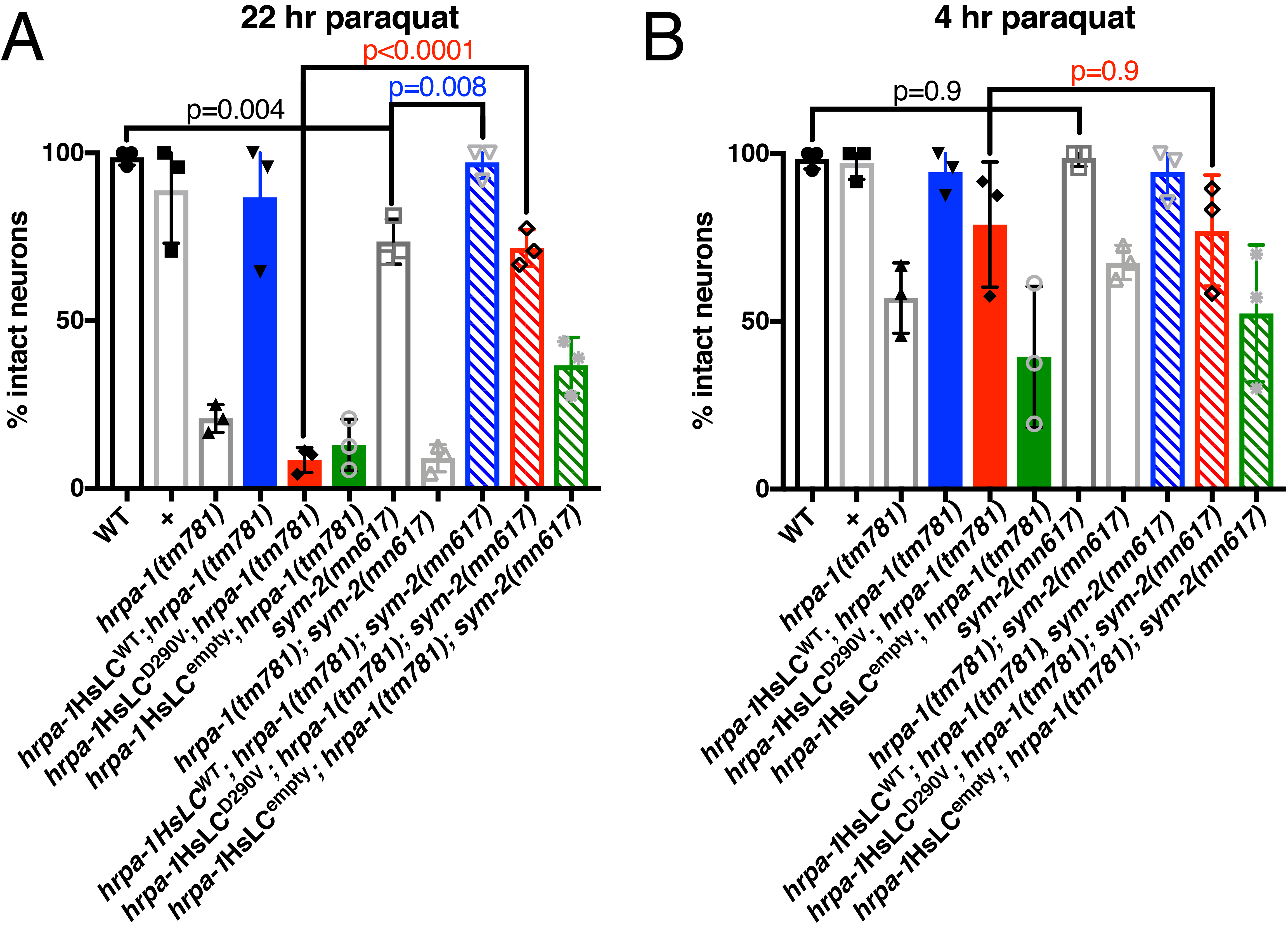
sym-2 encodes an ortholog of the RNA-binding proteins hnRNPF and hnRNPH. A putative loss-of-function allele, sym-2(mn617) from (Davies et al., 1999) is a Y163N missense mutation, which lies N-terminal to the first RNA-recognition motif of SYM-2. Homozygous animals have no overt defects but animals heterozygous for sym-2(mn617) and homozygous for mec-8 mutations are embryonic lethal (Davies et al., 1999; Yochem et al., 2004). Additionally, sym-2(mn617) suppresses the exhaustion-induced locomotion defect of smn-1(cb131) animals (Walsh et al., 2020). We have previously shown that human hnRNPA2, an ortholog of the protein encoded by C. elegans hrpa-1, interacts with hnRNPF in vitro (Ryan et al., 2020). As the low complexity (LC) domains of hnRNPA2 and HRPA-1 are not well conserved, we replaced the third coding exon of the C. elegans hrpa-1 gene with the corresponding human protein sequence codon optimized for C. elegans expression, resulting in a chimeric HRPA-1 protein with the human LC domain (HRPA-1HsLCWT) (Ryan et al., 2020). A mutation in hnRNPA2, D290V, is associated with multisystem proteinopathy, a disease that causes degeneration of muscles, bone, and neurons (Kim et al., 2013). Expression of the mutant chimeric version of hrpa-1, hrpa-1HsLCD290V, causes glutamatergic neurodegeneration in phasmid neurons after oxidative stress in animals lacking endogenous hrpa-1 function (Ryan et al., 2020). To control for defects associated with array integration, we also created an empty array, hrpa-1HsLCempty, which contains no hrpa-1. We hypothesized that sym-2(mn617) might rescue hrpa-1HsLCD290V stress-induced neurodegeneration.
To test this hypothesis, we generated animals carrying sym-2(mn617) with hrpa-1(tm781) and hrpa-1HsLC variants. We first exposed all genotypes to 22 hours of paraquat-induced oxidative stress and counted intact neurons after dye filling. We found that sym-2(mn617) rescued hrpa-1HsLCD290V stress-induced neurodegeneration but we also found that about 25% of the neurons in sym-2(mn617) animals failed to dye fill after 22 hours of stress. Virtually all sym-2(mn617) neurons dye filled after only 4 hours of stress; therefore, the dye filling defect is degeneration induced by stress. Interestingly, we found that sym-2(mn617) neurodegeneration is sensitive to hrpa-1 function. Animals expressing both sym-2(mn617) and hrpa-1HsLCWT showed significant rescue of the sym-2(mn617) defect back to WT levels. hrpa-1(tm781), hrpa-1HsLCD290V, hrpa-1HsLCempty did not rescue the sym-2(mn617) defect.
The identification of neurodegeneration in sym-2(mn617) animals after longer exposure to oxidative stress was surprising, especially given the suppression of neuromuscular defects in smn-1(cb131) (Walsh et al., 2020) and improvement of hrpa-1HsLCD290V defects. However, HNRNPF, an ortholog of sym-2, is associated with the neurodevelopmental disorder Rett syndrome (Newnham et al., 2010), suggesting that HNRNPF has an important role in preserving neuronal integrity. Interestingly, although hrpa-1HsLCWT can rescue the sym-2(mn617) neurodegeneration, hrpa-1HsLCD290V has similar levels of neurodegeneration as sym-2(mn617), with substantially less neurodegeneration than observed for hrpa-1HsLCD290V alone. As such, sym-2 may be in a genetic pathway upstream of hrpa-1, although more work is needed to elucidate what this pathway is and how it leads to neurodegeneration.
Methods
Request a detailed protocolGlutamatergic neurodegeneration: Day 1 adult animals were washed off plates with M9 and incubated with DiD (Fisher DilC18(5) D307) in a microfuge tube as in (Perkins et al., 1986). After 1.5 hours, animals were spun down at 10000 rpm for 30 seconds and transferred to a regular NGM plate. After 30 minutes, animals were mounted on 2% (vol/vol) agar pads and immobilized in 30 mg/mL 2-3-butaneione monoxime (BDM, Sigma) in M9 buffer. Fluorescent neuronal cell bodies were visualized and scored for dye uptake under 63x objectives. There are 4 phasmid neurons per animal, two per side. Neurons were scored as intact if the cell body took up fluorescent dye. For trials with paraquat stress, animals were exposed to 2.5 mM paraquat on plates for the indicated time frame.
Statistical analysis: Data collection and analysis were performed by experimenters blinded to genotype. Quantitative data were analyzed using GraphPad Prism 7. Two tailed t-test was used to determine significance for neurodegeneration assays. A value of P < 0.05 was used to establish statistical significance. Error bars in figures represent error of the mean (S.E.M.). A one-way ANOVA was performed with a Tukey post-hoc test for the 55 comparisons; these corrected p values are presented in the figure legend.
Reagents
C. elegans were maintained on Nematode Growth Media (NGM) that was seeded with E. coli strain OP50 as a food source according to established protocols (Brenner, 1974). Animals were maintained at 20˚C. All animals except sym-2(mn617) and N2 were maintained over the tmC25[tmIs1241] balancer. Experimenter was blinded to genotype for all trials and all trials were independent.
The sym-2 mutant strains described herein are available by request (anne_hart@brown.edu). They are:
| HA3578 | sym-2(mn617) II |
| HA3610 | hrpa-1(tm781)/tmC25[tmIs1241] IV; sym-2(mn617) II |
| HA3766 | rtIs83[hrpa-1p::hrpa-1HsLCWT::hrpa-1 3’UTR + elt-2p::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV; sym-2(mn617) II |
| HA3768 | rtIs82[hrpa-1p::hrpa-1HsLCD290V2::hrpa-1 3’UTR + elt-2p::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV; sym-2(mn617) II |
| HA3769 | rtIs93[hrpa-1p::hrpa-1HsLCempty1::hrpa-1 3’UTR + elt-2::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV; sym-2(mn617) II |
The following previously published strains were used in this study:
N2 Wild type, Bristol, (Brenner, 1974)
From (Ryan et al., 2020):
| HA3608 | tmC25[tmIs1241]/+ IV |
| HA3450 | hrpa-1(tm781)/tmC25[tmIs1241] IV |
| HA3655 | rtIs83[hrpa-1p::hrpa-1HsLCWT::hrpa-1 3’UTR + elt-2p::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV |
| HA3659 | rtIs82[hrpa-1p::hrpa-1HsLCD290V2::hrpa-1 3’UTR + elt-2p::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV |
| HA3708 | rtIs93[hrpa-1p::hrpa-1HsLCempty1::hrpa-1 3’UTR + elt-2::GFP + salmon sperm DNA]; hrpa-1(tm781)/tmC25[tmIs1241] IV |
All strains used here are available by request (anne_hart@brown.edu).
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Some strains were provided by the National Bioresource Project at the Tokyo Women’s Medical University School of Medicine funded by the Ministry of Education.
References
Funding
VHR was supported by Grant F31NS110301 from NINDS, National Institutes of Health. Other support from NIH-NINDS P01 N5066888 (to ACH) and Judith & Jean Pape Adams Charitable Foundation.
Reviewed By
AnonymousHistory
Received: January 4, 2021Revision received: January 21, 2021
Accepted: February 1, 2021
Published: February 3, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ryan, VH; Hart, AC (2021). sym-2 loss-of-function causes glutamatergic neurodegeneration after oxidative stress. microPublication Biology. 10.17912/micropub.biology.000363.Download: RIS BibTeX