Department of Microbiology, University of Georgia, Athens, GA, USA 30602
Department of Molecular and Cellular Pharmacology, University of Miami Miller School of Medicine, Miami, FL, USA 33101
Abstract
The autophagy-related protein Atg27p has been previously shown to localize to the autophagy-specific pre-autophagosomal structure (PAS) as well as to several organelles, including the late Golgi, the vacuolar membrane, and the endosome. Given that Atg27p localization to the vacuolar membrane in particular has been shown to be dependent on both its C-terminal tyrosine sorting motif and the AP-3 adaptor, and that Atg27p can be found in clathrin-coated vesicles, we set out to determine whether Atg27p localization inside cells is dependent on clathrin or on any of its cargo adaptors. We report that Atg27p localization is clathrin- and Ent3p/5p-dependent.
Description
The autophagy-related protein Atg27p has been previously shown to localize to the autophagy-specific pre-autophagosomal structure (PAS) as well as to several organelles, including the late Golgi or trans-Golgi network (TGN), the vacuolar membrane, and the early and late endosomes (Segarra et al. 2015). Moreover, Atg27p localization to the vacuolar membrane, in particular, is dependent on both its C-terminal tyrosine sorting motif and the AP-3 adaptor (Segarra et al. 2015; Suzuki and Emr 2019), but not other yeast AP-related adaptors (Segarra et al. 2015). We recently reported that Atg27p can be found in clathrin-coated vesicles (CCVs; Ding, Segarra et al. 2016, See accompanying micropublication). For this reason, we set out to determine whether Atg27p localization is dependent on clathrin or on any other of its cargo adaptors.
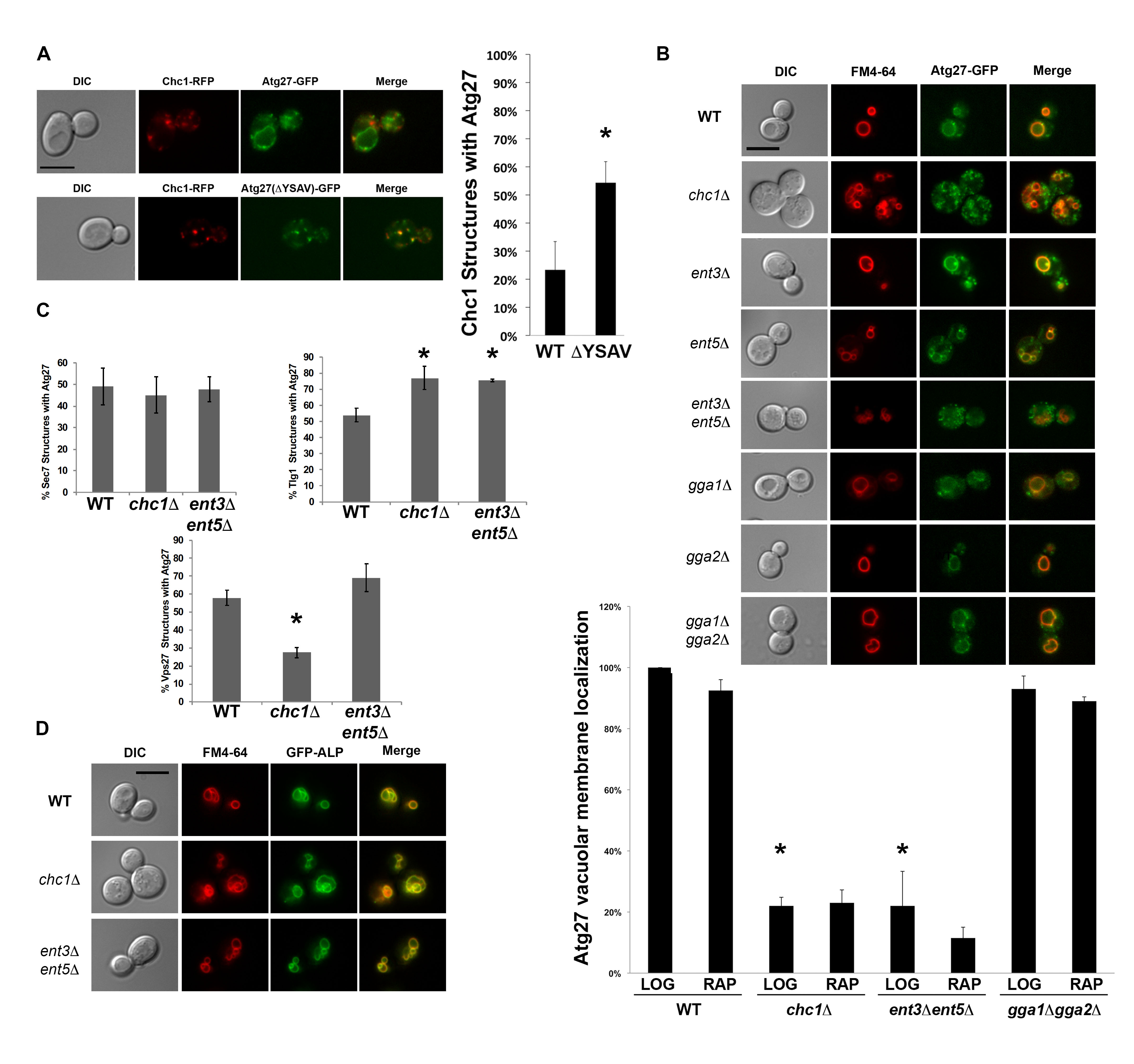
We found that Atg27p co-localizes with Chc1p (Figure 1A), even in the absence of its C-terminal tyrosine sorting motif YSAV (ΔYSAV). Thus, the presence of Atg27p in CCVs does not seem to require the anterograde transport of Atg27p to the vacuolar membrane. Atg27p mutant molecules deleted for their YSAV motif display increased colocalization with clathrin (Figure 1A). AP-3 sorting to the vacuole of these ΔYSAV mutants is most likely impaired, leading to increased amounts of Atg27p entering the traditional TGN/endosomal pathway.
To examine the overall localization pattern of Atg27p in cells lacking clathrin, we imaged cells expressing GFP-tagged Atg27p that were deleted for CHC1. In chc1∆, Atg27p localizes to small punctate structures throughout the cell and its vacuolar membrane localization is lost (Figure 1B). This was surprising because Atg27p has been shown to be an AP-3 cargo (Segarra et al. 2015) and the trafficking of AP-3 cargo, such as alkaline phosphatase (ALP; Stepp et al. 1997; Cowles et al. 1997), has traditionally been considered to be clathrin-independent (Vowels and Payne 1998). Similarly, we examined vacuolar membrane localization of Atg27p in cells missing the other TGN/endosomal clathrin adaptors, epsins or ggas. Double deletion of the ENT3 and ENT5 adaptor genes appear to recapitulate the vacuolar membrane localization defect seen in the clathrin-null cells, suggesting that transport of Atg27p is also dependent on the Ent3p and Ent5p adaptor proteins (Figure 1B). Ent5p was also identified in the mass spectrometry screen for CCV components, similar to Atg27p (WT screen, Ding, Segarra et al. 2016). The Ent3p and Ent5p epsin-like proteins have been shown previously to have separate roles in clathrin-mediated TGN/endosomal traffic (Costaguta, Duncan et al. 2006). Our finding that the Atg27p localization phenotype in clathrin null cells can only be recapitulated by combined deletion of ENT3 and ENT5 may indicate that clathrin is acting at more than one step in Atg27p TGN/endosomal transport. It is also interesting that the vacuolar membrane localization of Atg27p is not affected in any of the GGA adaptor mutants, as GGA clathrin adaptors were shown by Casler and Glick to be required for traffic of vacuolar cargo from the maturing Golgi (2020).
To determine the cellular location of the small Atg27p puncta in the chc1∆ and ent3∆ ent5∆ mutants, we quantified their co-localization with traditional Golgi/endosomal markers Sec7p, Tlg1p, and Vps27p. Atg27p puncta in the clathrin-null and the ent3∆ ent5∆ double deletion mutants partially localize to the Sec7p- and Tlg1p-marked TGN/recycling endosome and the Vps27p-marked pre-vacuolar endosome (Figure 1C). The budding yeast TGN has been shown to serve as the cell’s early/recycling endosome (Day et al. 2018), potentially explaining why certain organellar markers such as Sec7p and Tlg1p are sometimes reported to co-localize (Grissom et al. 2020). All in all, we can conclude that the Atg27p puncta in the clathrin null and in the ent3∆ ent5∆ cells partially localize to TGN/recycling endosome and the pre-vacuolar endosome.
The observed loss of vacuolar membrane localization was specific to Atg27p and not to all AP-3 cargoes, since ALP was able to localize to the vacuolar membrane in the clathrin and ent3 ent5 mutants (Figure 1D). This suggests that Atg27p is a non-canonical AP-3 cargo in that its vacuolar membrane localization is both AP-3 and clathrin- and epsin-dependent. Recent studies showed that Atg27p is recycled from the vacuolar membrane to endosomes and then the TGN (Suzuki and Emr 2018). Possibly, in the absence of clathrin or the epsins, Atg27p recycling to the TGN is impaired, leading to the loss of Atg27p from the vacuolar membrane and accumulation in TGN/endosomal compartments. Of interest, in chc1∆ cells, Atg27p was decreased on Vps27p-positive structures (Figure 1C), suggesting that clathrin’s major role is in a later retrieval step from the TGN.
Methods
Request a detailed protocolYeast and plasmid methods
Standard methods and media were used for genetic manipulations, growth, and transformation of yeast (Guthrie and Fink 1991). To induce autophagy, log phase cells were treated with rapamycin (LC Laboratories, R‐5000) at 0.2 µg/mL for at least 2 hours at 30°C. Saccharomyces cerevisiae strains used in this study are listed in the table below. Unless otherwise indicated, the Longtine method was used for yeast construction (Longtine et al. 1998). pGFP-ALP was used as an ALP localization reporter (Cowles et al. 1997).
Yeast strains used in this study
| Name | Alias | Genotype | Panel | Reference |
| SL5970 | Chc1-RFP Atg27-GFP | MATa leu2 ura3 trp1 his3 lys2 CHC1-RFP::KANMX6 ATG27-GFP::HISMX | 1A
|
This Paper |
| SL6154 | Chc1-RFP Atg27(ΔYSAV)-GFP | MATa leu2 ura3 trp1 his3 lys2 CHC1-RFP::KANMX6 ATG27(-YSAV)-GFP::HISMX | ||
| SL5837 | Atg27-GFP | MATα leu2 ura3-52 trp1 his3-∆200 ATG27-GFP::HISMX6 | 1B
|
Segarra
et al. 2015 |
| SL6240 | chc1Δ Atg27-GFP | MATa leu2 ura3-52 trp1 his3-∆200 chc1∆::LEU2 ATG27-GFP::HISMX | This paper
|
|
| SL5945 | ent3Δ Atg27-GFP | MATa leu2 ura3-52 trp1 his3-∆200 ent3Δ::TRP1 ATG27-GFP::HISMX6 | ||
| SL5946 | ent5Δ Atg27-GFP | MATa leu2 ura3-52 trp1 his3-∆200 ent5Δ::TRP1 ATG27-GFP::HISMX6 | ||
| SL5947 | ent3Δ ent5Δ Atg27-GFP | MATα leu2 ura3-52 trp1 his3-∆200 ent3Δ::TRP1 ent5Δ::TRP1 ATG27-GFP::HISMX6 | ||
| SL6214 | gga1Δ Atg27-GFP | MATα leu2 ura3-52 trp his3-∆200 lys2-801 gga1Δ::HIS3 ATG27-GFP::HISMX | ||
| SL6215 | gga2Δ Atg27-GFP | MATα leu2 ura3-52 trp his3-∆200 lys2-801 gga2Δ::TRP1 ATG27-GFP::HISMX | ||
| SL6222 | gga1Δ gga2Δ Atg27-GFP | MATα leu2 ura3-52 trp his3-∆200 lys2-801 gga1Δ::HIS3 gga2Δ::TRP1 ATG27-GFP::HISMX | ||
| SL7575
|
chc1Δ Atg27-GFP Sec7-dsRed | MATa leu2 ura3-52 trp1 his3-∆200 scd1-v chc1∆::LEU2 ATG27-GFP::HISMX6 pSEC7-dsRed (316) | 1C
|
This paper |
| SL7576
|
chc1Δ Atg27-GFP
mcherry-Tlg1 |
MATa leu2 ura3-52 trp1 his3-∆200 scd1-v chc1∆::LEU2 ATG27-GFP::HISMX6 pRS316-mcherry-TLG1 | ||
| SL7577
|
chc1Δ Atg27-GFP Vps27-RFP | MATa leu2 ura3-52 trp1 his3- ∆ 200 scd1-v chc1∆::LEU2 ATG27-GFP::HISMX6 pRS316-VPS27-RFP | ||
| SL7572
|
ent3∆ ent5∆ Atg27-GFP
Sec7-dsRed |
MATα ura3-52 his3 ∆ -200 trp1901 leu2-3,112 lys2-801 suc2- ∆9 ent3∆::TRP1 ent5∆::TRP1 ATG27-GFP::HISMX6 pSEC7-dsRed (316) | ||
| SL7573
|
ent3∆ ent5∆ Atg27GFP
mcherry-TLG1 |
MATα ura3-52 his3 ∆ -200 trp1901 leu2-3,112 lys2-801 suc2- ∆9 ent3∆::TRP1 ent5∆::TRP1 ATG27-GFP::HISMX6 pRS316-mcherry-TLG1 | 1C
|
This paper |
| SL7574
|
ent3∆ ent5∆ Atg27GFP
Vps27-RFP |
MATα ura3-52 his3 ∆ -200 trp1901 leu2-3,112 lys2-801 suc2- ∆9 ent3∆::TRP1 ent5∆::TRP1 ATG27-GFP::HISMX6 pRS316-VPS27-RFP | ||
| SL7575
|
chc1Δ Atg27-GFP
Sec7-dsRed |
MATa leu2 ura3-52 trp1 his3- ∆200 scd1-v chc1∆::LEU2 ATG27-GFP::HISMX6 pSEC7-dsRed (316) | ||
| SL7576
|
chc1Δ Atg27-GFP
mcherry-Tlg1 |
MATa leu2 ura3-52 trp1 his3- ∆ 200 scd1-v GAL2? chc1∆::LEU2 ATG27-GFP::HISMX6 pRS316-mcherry-TLG1 | ||
| SL7577
|
chc1Δ Atg27-GFP
Vps27-RFP |
MATa leu2 ura3-52 trp1 his3- ∆ 200 scd1-v GAL2? chc1∆::LEU2 ATG27-GFP::HISMX6 pRS316-VPS27-RFP | ||
| SL6679 | pGFP-ALP (SL6610) | MATα leu2 ura3-52 trp1 his3-∆200 pGFP-ALP | 1D
|
This paper
|
| SL6681 | chc1Δ GFP-ALP | MATα leu2 ura3-52 trp1 his3-∆200 chc1∆::LEU2 pGFP-ALP | ||
| SL6680 | ent3Δ ent5Δ GFP-ALP | MATα leu2 ura3-52 trp1 his3-∆200 ent3Δ::TRP1 ent5Δ::TRP1 pGFP-ALP |
Microscopy methods
Vacuolar membrane staining with FM4-64 was carried out as described previously (Segarra et al. 2015). Microscopy was performed on an Olympus fluorescence BX61 upright microscope equipped with Nomarski differential interference contrast (DIC) optics, a Uplan S Apo 100x objective (NA 1.4), a Roper Cool-Snap HQ camera, and Sutter Lambda 10–2 excitation and emission filter wheels, and a 175 watt Xenon remote sourcewith liquid light guide. Image capture was automated using Intelligent Imaging Innovations Slidebook 4.01 for Mac. A series of optical Z-sections (0.25 μm) was captured for each cell analyzed. Prior to analysis, the stacks were deconvolved using the nearest neighbor algorithm. Representative single-plane micrographs from cells at log phase were chosen to be included in the figures.
To quantify co-localization of Atg27p with Chc1p or organellar markers, deconvolved Z-stacks were examined to confirm that both fluorescent signals were in the same plane, and that peak fluorescence overlapped in corresponding sections. Co-localization was expressed as the percent of structures of interest that contained the GFP-tagged Atg27p construct. To determine statistical significance, two-tailed Student’s t-tests were performed to compare each condition of interest to the WT control.
Acknowledgments
VAS thanks the Department of Biology and the Wanek School of Natural Sciences at High Point University for resources that allowed for the writing of this manuscript. We thank Greg Odorizzi for the GFP-ALP and SEC7-dsRed plasmids. We also thank David Katzmann for the mcherry-TLG1 and VPS27-RFP plasmids.
References
Funding
This work was supported by National Institutes of Health grant R01-GM055796 to SKL and T32-HL07188 to VAS.
Reviewed By
AnonymousHistory
Received: January 16, 2021Revision received: March 21, 2021
Accepted: March 24, 2021
Published: March 29, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Segarra, VA; Sharma, A; Lemmon, SK (2021). Atg27p localization is clathrin- and Ent3p/5p-dependent. microPublication Biology. 10.17912/micropub.biology.000381.Download: RIS BibTeX