Abstract
unc-11 is the only C. elegans ortholog of mammalian PICALM/AP180, paralogs that play an important role in Clathrin-Mediated Endocytosis (CME) and the recycling of a subset of SNAREs, the vesicle-associated membrane proteins (VAMPs). In this publication we report the creation of a new unc-11 allele that is endogenously-tagged with GFP just upstream of the stop codon. Moreover, we demonstrate that the UNC-11::GFP fusion protein functions like wild type with an expression pattern similar to UNC-11 antibody staining described previously.
Description
UNC-11 is an ortholog of mammalian PICALM (phosphatidylinositol binding clathrin assembly protein) and its neuron-specific paralog, AP180 (Maritzen et al. 2012). PICALM facilitates recycling of Vesicle Associated Membrane Proteins (VAMPs) from the plasma membrane (Miller et al. 2011, Sahlender et al. 2013 and Miller et al. 2015) and efficient clathrin-coated endocytosis (Huang et al. 2004, Miller et al. 2015 and Ishikawa et al. 2014). unc-11 function in C. elegans is likely conserved as SNB-2 (VAMP2) and the glutamate receptor, GLR-1, both accumulate at the plasma membrane in unc-11(e47) null mutants (Nonet et al. 1999, Burbea et al. 2002). Mutant phenotypes outside the worm nervous system have not been described.
To determine the endogenous expression pattern and subcellular localization of unc-11, we created a functional GFP knock-in at the endogenous unc-11 locus using the CRIPSR approach outlined by Dickinson et al. 2015. Specifically, a GFP::AID::3xFLAG coding sequence interrupted by four synthetic introns was inserted just 5′ to the unc-11 stop codon. Successful knock-in was verified by sequencing the insertion junctions and expression of GFP.
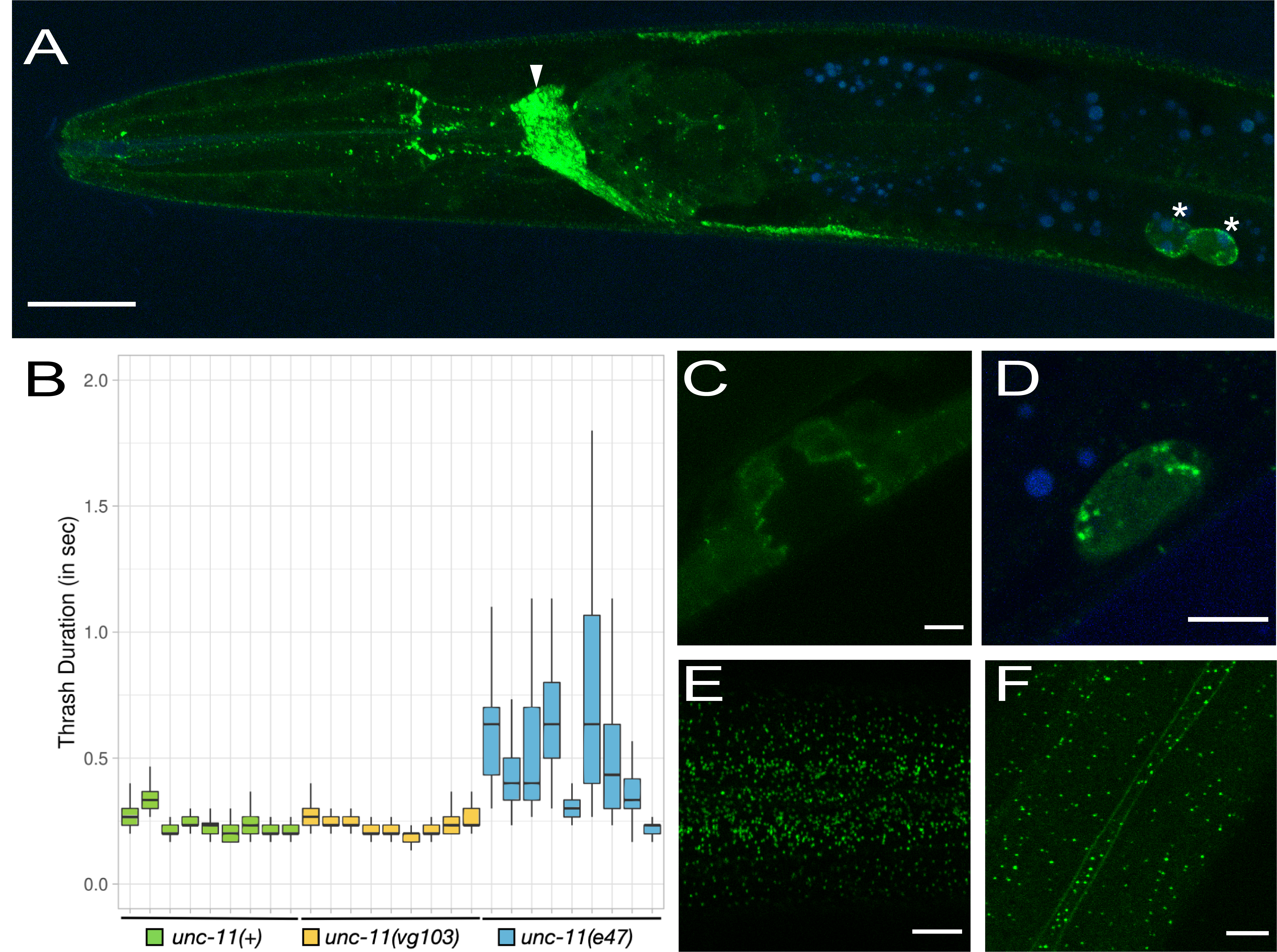
To ask if the addition of GFP at the C-terminus of UNC-11 compromises unc-11 function we performed a thrashing assay to examine the general health of the nervous system (Figure 1B). We compared the thrash duration (see methods) of unc-11(+) with the CRISPR knock-in strain, unc-11(vg103) and the null mutant, unc-11(e47). We found no obvious differences between unc-11(+) and unc-11(vg103). Average thrash duration during a 45 second interval was 0.251 seconds (n=9, Std. Dev. = 0.042) for unc-11(+) and 0.248 seconds (n=9, Std. Dev. = 0.031) for unc-11(vg103). By contrast, the average thrash duration for unc-11(e47) was significantly higher at 0.566 seconds with a striking increase in variability (n=9, Std. Dev. = 0.196).
In general, UNC-11::GFP expression matched that of UNC-11 antibody staining described previously (Nonet et al. 1999) with some noted exceptions. Similar to UNC-11 antibody staining, UNC-11::GFP expression appeared pan-neuronal and was also observed within coelomocytes (Figure 1A and 1D). Not described by Nonet el al 1999, we observed GFP puncta in apical epithelial tissue just below the cuticle surface in both the head and in the body (Figure 1E and 1F), an accumulation of UNC-11::GFP at the plasma membrane in the seam cells (Figure 1F) and weak staining in the vulval precursor cells (VPCs) at the interface between the VPCs and the vulval lumen (Figure 1C). Finally, Nonet et al. described diffuse staining within the intestine whereas we did not observe detectable levels of intestinal expression, punctate or otherwise (Figure 1A).
Plasmid construction was done in the context of a course entitled Advanced Molecular Techniques at California State University, East Bay. This strain will be made available at the CGC. Given the inclusion of an AID tag and GFP, this strain will be useful for cell-specific protein degradation using auxin-inducible (AID) or GFP nanobody::ZIF-1 degradation methods (Wang et al. 2007 and Zhang et al. 2015).
Methods
Request a detailed protocolThrashing Assay: Individual worms were placed on a glass slide with a 3% agar pad and about 75 ul of M9 buffer. Slides were then mounted onto a DIY microscope (original design by Kenji Yoshino) and videos were captured at 30 frames per second using an iPhone 7. To open the images in FIJI, the .MOV files were converted to an uncompressed AVI format using ff.WORKS software. The duration of each thrash was measured in FIJI by logging each frame number at which the worm completed a single body bend in one direction. Thrash duration (in seconds) was then calculated as the number of frames per body bend divided by 30 frames per second. Approximately, 1450 frames (~45 seconds) of video were reviewed for each animal. Body bends that did not fall below 150 degrees were not counted as a true body bend. When a body bend angle was suspected to be greater than 150 degrees, the angle ruler in Fiji (Image J) was used. Brief intervals where thrashing occurred along the Z-axis or the worm left the field of view were not included. The data was graphed using Plots of Differences available at https://huygens.science.uva.nl/PlotsOfDifferences/ (Joachim Goedhart, 2019).
Confocal Microscopy: unc-11(vg103) animals were anesthetized with sodium azide (10 mM) and imaged using a Leica SP8 confocal. To visually distinguish true GFP signal from intestinal autofluorescence for Figures 1A and 1D, Z-stacks were captured using a sequential scanning protocol: Seq 1: The 488 laser was set at 2.4%. PMT1 (green) collected wavelengths from 494 to 544 nm (capturing GFP and autofluorescence) while PMT2 (blue) collected wavelengths from 650 to 798 nm (autofluorescence only). Seq 2: The 552 laser was set at 1.0 % and PMT1 (blue) collected wavelengths from 598 to 709 nm (autofluorescence only). Adjustments were made to brightness and contrast of the blue channels to bring out the autofluorescence then the green and blue channels were combined and relevant focal planes were subject to maximum projection. The other images did not emit detectable autofluorescence and so the blue channel images were not included in the Z projection.
Reagents
The sgRNA/Cas9 plasmid (pDQ1022) was created by Q5 SDM (NEB) using pJW1219 as template (a plasmid containing sgRNA(F+E) and Peft-3::Cas9; Addgene # 61250) and the following primer pair: CGACTAGACTATAATCCAAAgtttaagagctatgctggaa and caagacatctcgcaatagga. The unc-11 target sequence is in all caps and the position of the unc-11 STOP codon is in bold. The GFP::SEC repair template (pDQ1023) was created by Gibson Assembly of pJW1583 (A GFP^SEC^AID*::3xFLAG vector with ccdB sites for cloning homology arms; Addgene # 121054) digested with AvrII and SpeI and two unc-11 homology arms (HA) amplified by PCR (Q5 DNA polymerase – NEB). Primers used to amplify the 5’HA and 3’HA homology arms from wild type genomic DNA include: ACGACGGCCAGTCGCCGGCATGCGATGGGTTAGTTTATCG and CCTGAGGCTCCCGATGCTCCTAATCCAAATGGATCGGCTG (5’HA); AGGATGACGATGACAAGAGATAGTCTAGTCGTCTATTTCTC and TATGACCATTTATCGATTTCGCAATAGAGATAAAGTTGCA (3’HA). unc-11(vg103[unc-11::gfp]) (DQ3188) was created as described by Dickinson et al. 2015. Successful knock-in was verified by PCR amplification and sequencing of the junction fragments and by GFP expression.
Acknowledgments
We thank Dr. Jordan Ward for technical advice during course development and Dr. Noelle L’Etoile for advice during manuscript preparation.
References
Funding
The Department of Biology paid for the reagents used in the lab course, Biol 622.
Reviewed By
Diana ChuHistory
Received: April 7, 2021Revision received: April 19, 2021
Accepted: April 19, 2021
Published: April 28, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Gallegos, M; Hermes, A; Patra, D (2021). Creation of a functional unc-11/PICALM GFP knock-in by CRISPR. microPublication Biology. 10.17912/micropub.biology.000389.Download: RIS BibTeX