Max Delbrück Center for Molecular Medicine in the Helmholtz Association, 13125 Berlin, Germany
Abstract
We recently identified FAcilitates Chromatin Transcription (FACT) as a reprogramming barrier of transcription factor (TF) mediated conversion of germ cells into neurons in C. elegans. FACT is a conserved heterodimer consisting of SPT16 and SSRP1 in mammals. Duplication events during evolution in C. elegans generated two SSRP1 homologs named HMG-3 and HMG-4, while SPT-16 is the only homolog of SPT16. Yet, the pseudogene F55A3.7 has nearly complete nucleotide sequence homology to the spt-16 gene. However, F55A3.7 lacks some spt-16 exons and DNA pieces so we named it sspt-16 (short spt-16). Surprisingly, the deletion mutant ok1829, which affects only the sspt-16 pseudogene, shows similar germ cell reprogramming effects as described previously for FACT-depleted animals. We examined whether lack of sspt-16 affects other genes or chromatin accessibility, which may explain the permissiveness for germ cell reprogramming.
Description
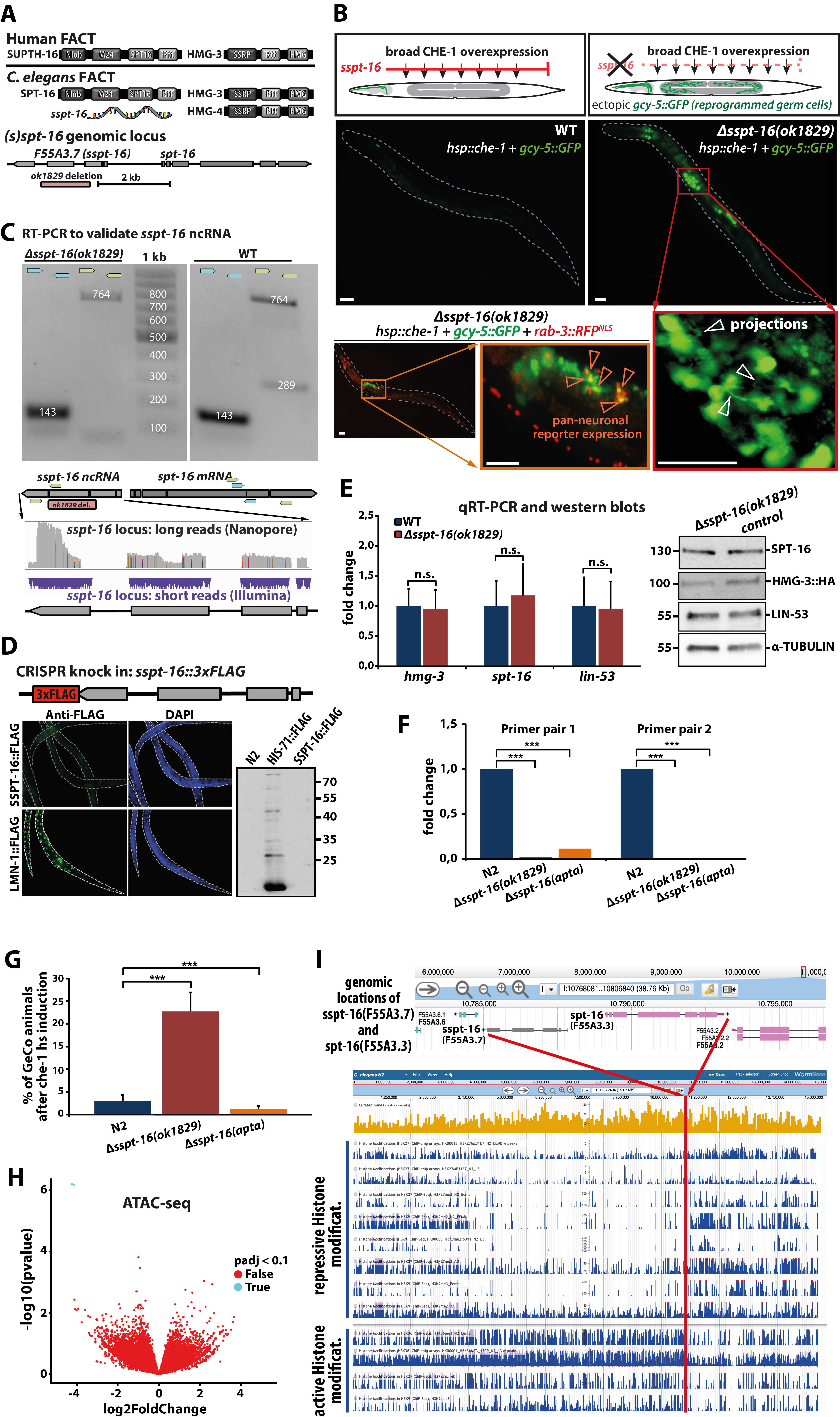
FAcilitates Chromatin Transcription (FACT) is a histone chaperone complex, which is implicated in a variety of biological processes by regulating nucleosome deposition and chromatin accessibility (Orphanides et al. 1998; Hammond et al. 2017). In C. elegans, we previously demonstrated that FACT is a reprogramming barrier of transcription factor (TF) mediated reprogramming to neuron-like cells by safeguarding intestinal and germ cell identities (Kolundzic et al. 2018). Its role as a reprogramming barrier is conserved such that FACT depletion in human fibroblast culture enhanced reprogramming efficiency into both iPSCs and neurons (Kolundzic et al. 2018). FACT is a heterodimer consisting of two subunits: SUPT16H (suppressor of Ty 16 homolog, also known as SPT16) and SSRP1 (structure-specific recognition protein 1) in mammals (Orphanides et al. 1999). In C. elegans, SSRP1 has two homologs: HMG-3 and HMG-4, while SPT-16 is the single homolog of SPT-16 (Kolundzic et al. 2018). The existence of the two alternative FACT subunits HMG-3 and HMG-4 is the consequence of gene duplication, resulting in two functional paralogous proteins with distinct expression patterns: HMG-3 is expressed exclusively in the germline, while HMG-4 is expressed primarily in the soma (Kolundzic et al. 2018). Interestingly, SPT-16 also went through a gene duplication event, resulting in the uncharacterized pseudogene F55A3.7, which we named sspt-16 (short spt-16), because it lacks some genomic DNA pieces and an open reading frame when compared to the spt-16 gene locus (Figure 1A).
Pseudogenes can have coding-independent regulatory functions during differentiation and tumorigenesis (Singh et al. 2020). A well-studied example is the pluripotency TF and tumor marker factor OCT4 with respect to its seven pseudogenes, which are differentially expressed in different tumors (Poursani et al. 2016). For instance, in hepatocellular carcinoma, OCT4-pg4 (OCT4 pseudogene 4) functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 (Wang et al. 2013).
We wondered whether sspt-16 has a function which may be related to its origin gene spt-16 and FACT with regard to safeguarding germline identity in C. elegans. To address this question, we combined the deletion mutant ok1829, which lacks a large genomic fragment only of sspt-16 (Figure 1A), with a strain carrying the heat-shock inducible TF CHE-1 and the ASE neuron fate reporter gcy-5p::GFP. We demonstrated previously that overexpressed CHE-1 induces germ cell reprogramming to ASE neuron-like cells upon HMG-3 depletion (Kolundzic et al. 2018) and tested whether lack of the pseudogene sspt-16 causes a related germ cell reprogramming phenotype (Figure 1B). Indeed, after broad induction of the ASE neuron-specifying TF CHE-1 in L4 animals, ectopic expression of the ASE neuron fate reporter gcy-5p::GFP could be detected in the germline (assessed after 24 h) (Figure 1B). Some reprogrammed germ cells showed axo-dendritic projections (Figure 1B, bottom right), indicating that they acquired neuronal-like features. Additionally, reprogrammed germ cells also express the pan-neuronal marker rab-3p::NLS::tagRFP, corroborating the notion that the sspt-16 depletion background allows conversion of germ cells to neuron-like cells upon induction of CHE-1 overexpression. Taken together, these results indicate that the pseudogene sspt-16 exhibits a similar safeguarding function of germline identity as previously described for FACT (Kolundzic et al. 2018).
In order to understand how sspt-16 contributes to safeguarding the germline we proved that it is transcribed, which we could verify by detecting sspt-16 RNA transcripts through PCR using intron/exon border spanning primers (Figure 1C, top). Additionally, we detected sspt-16 RNA transcripts in long-read RNA-seq by Nanopore, confirming that the transcript underwent splicing events and consists of four exons (Figure 1C, bottom). The sspt-16 transcript should be considered as a non-coding RNA (ncRNA) because it contains two premature stop codons and a CRISPR-mediated FLAG epitope tag insertion at the presumptive C-terminus did not yield in any protein detection for immune staining and blotting (Figure 1D). Furthermore, inspection of polysome tracks provided in JBrowse (WS280) and a recent ribosome profiling study (Malone et al. 2017) do not indicate that a full-lenght sspt-16 (F55A3.7) peptide is generated. However, we cannot rule out the possibility of shorter peptides originating from the sspt-16 locus.
Since our lab has identified other reprogramming barriers in the past (Tursun et al. 2011; Kolundzic et al. 2018; Hajduskova et al. 2019), we wanted to assess whether the expression levels of known factors are altered in an sspt-16 depleted background, which could explain the resulting permissiveness for germ cell reprogramming in ok1829 animals. We checked the sspt-16(ok1829) deletion background for gene expression changes of the germline FACT members spt-16 and hmg-3, as well as for lin-53, another histone chaperone which blocks reprogramming of germ cells by promoting the formation of repressive chromatin barriers (Tursun et al. 2011). We performed gonad specific qPCR and whole worm western blots of WT compared to sspt-16(ok1829) animals, but could not detect any significant difference neither on mRNA nor on protein level for the tested factors (Figure 1E). This outcome, that lack of sspt-16 does not affect the neighboring spt-16 gene or other known reprogramming barrier factors, suggests that sspt-16 is required for safeguarding germline identity through other mechanisms.
To investigate the role of sspt-16 in safeguarding germ cells, we decided to examine whether it is sufficient to create permissiveness for germ cell reprogramming if only the sspt-16 transcript is eliminated but not the genomic DNA sequence as in the sspt-16(ok1829) mutant. We therefore knocked in an aptamer-ribozyme sequence at the 3’ end of WT sspt-16 using CRISPR/Cas. The cleavage activity of the ribozyme leads to RNA decay in absence of tetracycline (Wurmthaler et al. 2019). Testing by qPCR with two different primer pairs using WT animals as reference and sspt-16(ok1829) animals as positive control confirmed that sspt-16 transcripts were efficiently depleted in the new aptamer-ribozyme strain sspt-16(bar37), from now on referred as sspt-16(apta) (Figure 1F). However, in contrast to the deletion allele sspt-16(ok1829), where we can consistently see a germ cell conversion phenotype in almost 25% of animals, we couldn’t detect ectopic GFP expression in the germline of sspt-16(apta) animals. This finding suggests that it is not only the absence of sspt-16 ncRNA, but also the lack of genomic DNA of the sspt-16 locus, which is required to create permissiveness for germ cell reprogramming (Figure 1G).
We speculated that the presence of the sspt-16(ok1829) deletion could result in altered chromatin accessibility states in germ cells thus making them more amenable for CHE-1-mediated reprogramming to neurons as previously observed upon depletion of FACT (Kolundzic et al. 2018). To test this hypothesis, we performed a germline-specific Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq). However, we found no significant differences in chromatin accessibility of WT vs. sspt-16(ok1829) germlines (Figure 1H). Only two differentially accessible loci were identified with an adjusted p value of <1, one of which represents the ok1829 deletion itself (Figure 1H). As we could not find significant chromatin accessibility changes in sspt-16(ok1829) germlines, we speculate that differences at the level of chromatin modifications rather than its accessibility may be contributing to the decreased maintenance of germ cell identity in these mutants. This notion is supported by an interesting observation when revisiting publicly available data for active and repressive chromatin modifications (Figure 1I). When inspecting ChIP-seq and ChIP-chip tracks for repressive (H3K27me3 emb/L3/adult, H3K9me2/3 emb/L3/adult) and active (H3K36me3 emb/L3, H3K9ac L3, H3K77ac adult) H3 modifications, it appears that the sspt-16 / spt-16 locus lies within a region of the right arm of chromosome I, which delineates a boundary for repressive and active chromatin. Thus, we speculate that the lack of genomic DNA within the sspt-16 locus may lead to a boundary weakening, which could cause spreading of active chromatin and thereby allow activation of ectopic gene expression. Yet, the exact mechanism of how lack of the sspt-16 locus causes permissiveness for germ cell reprogramming remains to be determined and future investigation should focus on changes of chromatin modifications in the sspt-16 deletion mutant.
Methods
Request a detailed protocolC. elegans strains and maintenance
All strains used were grown in nematode growth medium (NGM) at 15°C as previously described (Brenner 1974). Heat-shocking of strains was performed at 37° C for 30 min following incubation at 25° C O/N.
RNA-seq
Library preparation for RNA sequencing was carried out using NEXTFLEX Rapid Directional RNA-Seq Kit 2.0 (Bioo Scientific) and SQK-DCS109/EXP-NBD104 (Nanopore) according to the manufacturer’s instructions. Libraries were sequenced using paired‐end sequencing length of a 100 nucleotides on a HiSeq 4000 machine (Illumina) and Minion (Nanopore).
Gonad dissection
Gonads were dissected as previously described (Jones et al. 1996).
Gonad specific ATAC-seq
Each replicate of dissected gonads (50 gonad arms in 20 µl ddH2O in a 1.5 ml Eppendorf tube) was filled up with 700 µl of freshly prepared and precooled buffer A (15 mM Tris–HCl pH 7.5/2 mM MgCl2/0.34 M sucrose/0.15 mM spermine/0.5 mM spermidine/1 mM DTT/0.5 mM PMSF/0.25% NP40/0.1% Triton X-100) (Ooi et al. 2010) and subjected to 30 strokes of a tight fitting plastic pestle to open up the gonads and to isolate the nuclei. Subsequently, the pestle was rinsed with 300 µl of buffer A, each sample was spun for 5 min (5000 g/4° C) and the supernatant was discarded. The in this way purified nuclei were tagmented using the Nextera DNA Flex Library Prep Kit (Illumina) according to the manufacturer’s instructions, adding additional PBS and Tween-20 to the tagmentation buffer as described by (Corces et al. 2017) and sequenced using paired-end-sequencing length of 75 nucleotides on a HiSeq4000 machine (Illumina).
Reagents
List of strains
| Name | Genotype | Availability |
| N2 | Caenorhabditis Elegans | CGC |
| RB1524 | F55A3.7(ok1829) I. | CGC |
| BAT28 | otIs305 [hsp-16.2p::che-1::3xHA, rol-6(su1006)], ntIs1 [gcy-5p::GFP, lin-15(+)] V. | upon request |
| BAT372 | F55A3.7(ok1829) I; otIs305 [hsp-16.2p::che-1::3xHA, rol-6(su1006)], ntIs1 [gcy-5p::GFP, lin-15(+)] V. | upon request |
| BAT748 | barEx308 [hsp-16.2::H3_S10E::2xFLAG, hsp-16.2::His-71_S10E::2xFLAG, myo-2p::mCherry], otIs305 [hsp-16.2p::che-1::3xHA, rol-6(su1006)], ntIs1 [gcy-5p::GFP, lin-15(+)] V. | upon request |
| BAT1468 | barIs165 [myo-3p::lmn-1T40I::2xFLAG::SL2::NLS::tagRFP] | upon request |
| BAT1594 | F55A3.7(bar23[F55A3.7::3xFLAG]) I. | upon request |
| BAT1560 | hmg-3(bar24[hmg-3::3xHA]) I. | upon request |
| BAT1749 | hmg-3(bar24[hmg-3::3xHA]) I; F55A3.7(ok1829) I. | upon request |
| BAT2099 | F55A3.7(bar37[F55A3.7::aptamer]) I; otIs305 [hsp-16.2p::che-1::3xHA, rol-6(su1006)], ntIs1 [gcy-5p::GFP, lin-15(+)] V. | upon request |
| BAT2192 | F55A3.7(ok1829) I; otIs355 [rab-3::NLS::TagRFP]; otIs305 [hsp-16.2p::che-1::3xHA, rol-6(su1006)], ntIs1 [gcy-5p::GFP, lin-15(+)] V. | upon request |
List of (q)RT-PCR primers
| Forward primer | Sequence | Reverse primer | Sequence | Target Gene |
| oBT504 | GTGTGGGACCTATCTAAGA | oBT223 | TTACTGTTGTCTCTCTACCAC | lin-53 |
| oBT823 | GGATCGTTGGAGGCTCATACT | oBT824 | TCATTATCAGATTCAGCATTCAGG | spt-16 and sspt-16 |
| oBT827 | CATTTTCGAGTTTGGGAAGG | oBT828 | CCATTGAAATAGTCGAGTTGT | hmg-3 |
| oBT839 | GGAGGCCCACACTAATGGATTTAGATACACATC | oBT840 | CTGGATTCTTGAGATGGAAGTGC | spt-16 |
| oBT863 | CTGCTGGACAGGAAGATTACG | oBT864 | CTCGGACATTCTCGAATGAAG | cdc-42 |
| oBT4011 | GCTGAACAACGGGAGAAGGA | oBT4012 | ACGCGAAAATTGGCTGTACAAA | sspt-16 |
List of antibodies
| Primary AB | Host & Clonality | Dilution | Company | Secondary AB |
| Anti-α-tubulin | mouse, mono | 1:10000 | Sigma | Anti-mouse-HRP, 1:10000, Santa Cruz Biotechnology Inc. |
| Anti-FLAG | mouse, mono | 1:1000 | Sigma | Anti-mouse-HRP, 1:10000, Santa Cruz Biotechnology Inc. or Anti-mouse AlexaFluor488, 1:1500, Molecular Probes |
| Anti-HA-HRP | chicken, poly | 1:1000 | Abcam | – |
| Anti-LIN-53 | rabbit, mono | 1:2000 | Pineda | Anti-rabbit-HRP, 1:10000, Santa Cruz Biotechnology Inc. |
| Anti-SPT-16 | guinea pig, mono | 1:2000 | Pineda | Anti-guinea pig-HRP, 1:10000, Santa Cruz Biotechnology Inc. |
Acknowledgments
We thank Sergej Herzog and Anne Krause for technical assistance. We also thank the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40OD010440), for providing strains for this study. All procedures conducted in this study were approved by the Berlin State Department for Health and Social (LaGeSo). The authors declare that they have no competing interests.
References
Funding
This work was funded by the ERC-StG-2014-637530 and the Max Delbrueck Center for Molecular Medicine in the Helmholtz Association.
Reviewed By
AnonymousHistory
Received: April 16, 2021Revision received: May 4, 2021
Accepted: May 5, 2021
Published: May 18, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ofenbauer, A; Kraus, CM; Tursun, B (2021). The C. elegans pseudogene sspt-16 (F55A3.7) is required to safeguard germ cells against reprogramming. microPublication Biology. 10.17912/micropub.biology.000392.Download: RIS BibTeX