Abstract
The FLP-1 neuropeptide is involved in locomotion, mechanosensation, and reproduction. Strain RB2126 is reported to contain the flp-1(ok2811) mutation, a prospective null allele. Here we report that strain RB2126, in addition to flp-1(ok2811), also contains two additional background mutations. One of the mutations gives the animals a constitutive dauer phenotype (Daf-c), and complementation testing confirms that it is in the Daf-c gene daf-11. The second mutation is isolated based on the fact that it partially suppresses the elevated body curvature of flp-1(ok2811) mutants, but it remains uncloned. The updated genotype of RB2126 strain is flp-1(ok2811); daf-11(yak155); yak156.
Description
The FLP family of neuropeptides contains at least 30 genes in C. elegans (Li and Kim, 2008). Some of the flp genes were found to play roles in locomotion, egg laying, and sleep, but most flp genes have unknown function (Li and Kim, 2008; Chang et al. 2015). flp-1 is one of the few flp genes whose mutations are reported to cause overt behavioral defects (Nelson et al. 1998; Hums et al. 2016; Buntschuh et al. 2018; Oranth et al. 2018). The initial flp-1 deletion alleles that were analyzed behaviorally (yn2 and yn4; Nelson et al. 1998) included a deletion of part of daf-10 (Ailion and Thomas, 2003; Buntschuh et al. 2018). This resulted in some of the initial behaviors that were attributed to FLP-1 peptides being caused by loss of daf-10 activity (Buntschuh et al. 2018). Subsequent analysis of newer flp-1 deletion alleles suggests that FLP-1 peptides are involved in nose touch sensitivity, locomotion, and reproduction (Buntschuh et al. 2018).
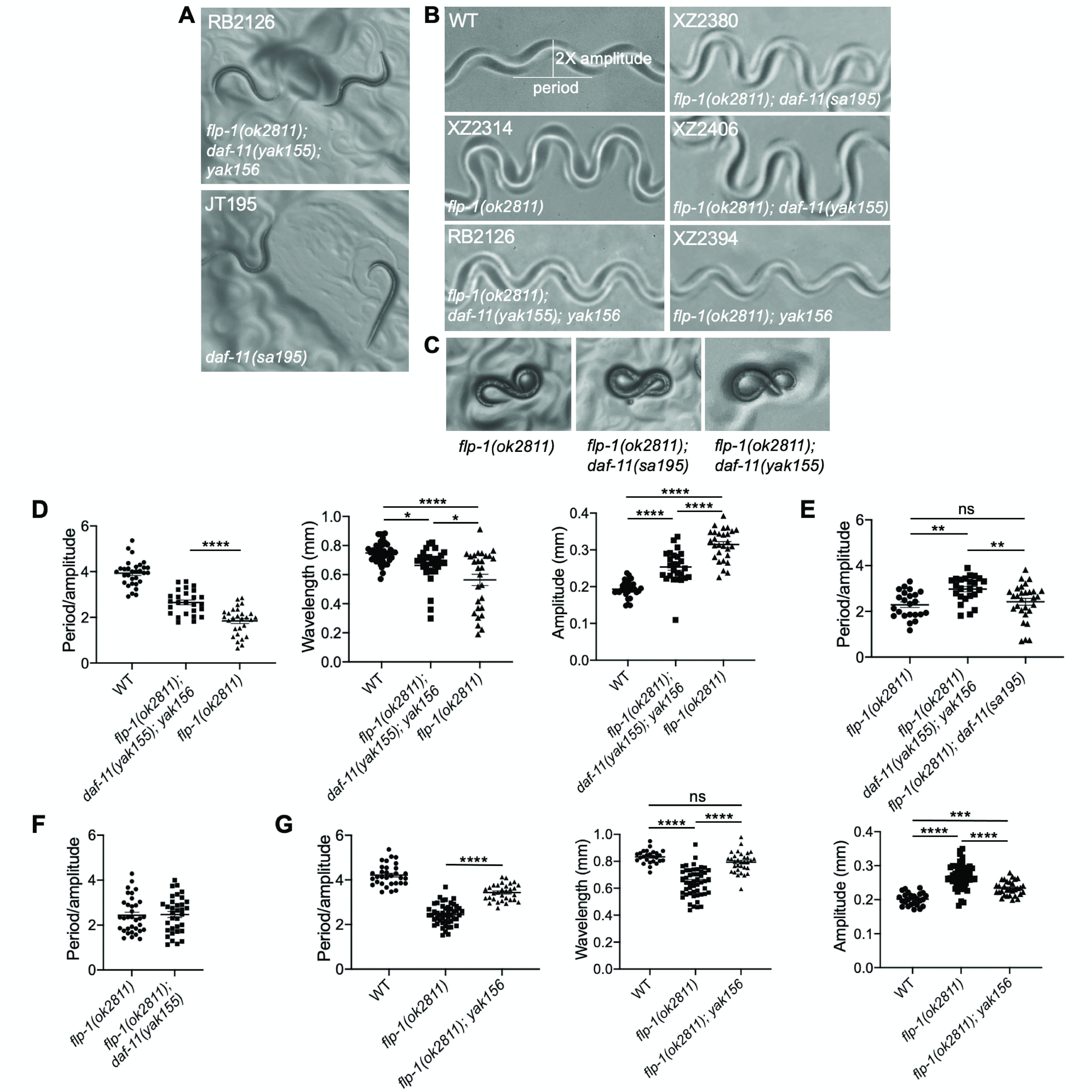
flp-1(ok2811) is a prospective null allele (Hums et al., 2016; Buntschuh et al.. 2018). We obtained strain RB2126 (Caenorhabditis Genetics Center, CGC) in order to analyze flp-1(ok2811) null mutant locomotion behavior. When the strain was grown at room temperature (23°C), some animals formed noticeable dauers similar to strains containing mutations in constitutive dauer (Daf-c) genes (Hu, 2007) (Fig. 1A). To examine whether the Daf-c phenotype was because of a background mutation, strain RB2126 was outcrossed three times. This outcross resulted in the segregation of two different mutants with readily apparent phenotypes: one resulting in deep body bends and a second in the Daf-c phenotype. PCR genotyping showed that the deep body bends co-segregated with the flp-1(ok2811) deletion. The mutation causing the Daf-c phenotype was mapped to chromosome V close to the daf-11 gene and a complementation test using daf-11(sa195) (a presumed null allele of daf-11) showed that it is in daf-11 (Birnby et al. 2000). These results suggest that flp-1(ok2811) mutants exhibit elevated body curvature, and that strain RB2126 has a background mutation in daf-11. This new daf-11 allele in strain RB2126 was named yak155.
flp-1(ok2811) mutants are known to exhibit elevated body curvature (Nelson et al. 1998; Chang et al. 2015; Hums et al. 2016, Buntschuh et al. 2018). Interestingly, strain RB2126 (flp-1(ok2811); daf-11(yak155)) had significantly reduced deep body bends in comparison to strain XZ2314 (flp-1(ok2811); three times outcrossed from strain RB2126) (Fig. 1B, 1D). This suggests that either the daf-11(yak155) mutation partially suppresses the phenotype of flp-1(ok2811) or that the RB2126 strain has other background mutation(s) that affect the body curvature. To test the possibility that the reduced deep body bends of flp-1(ok2811) in RB2126 strain are due to daf-11, double mutant strain flp-1(ok2811); daf-11(sa195) was constructed, and its body curvature was examined. flp-1(ok2811); daf-11(sa195) mutants (strain XZ2380) had similar body curvature as flp-1(ok2811) mutants (strain XZ2314) (Fig. 1B-1E). In addition, larvae (L2 to L3 stage) of flp-1(ok2811) and flp-1(ok2811); daf-11(sa195) often exhibit a characteristic body posture resembling the number ”8”, but this posture was not observed in RB2126 larvae (Fig. 1C). This result shows that daf-11(sa195) does not affect the elevated body curvature of flp-1(ok2811) mutants, suggesting that the body curvature of the RB2126 strain may be modified by another background mutation(s) other than daf-11(yak155).
To verify the presence of additional background mutation(s), the body curvature of flp-1(ok2811); daf-11(yak155) double mutants (strain XZ2406) and flp-1(ok2811) mutants (strain XZ2314) were compared. Body curvature in these strains was found to be similar, suggesting that strain RB2126 has at least one additional background mutation in an unknown gene that affects the body curvature of flp-1(ok2811) (Fig. 1C, 1F). In support of this, strain RB2126 was outcrossed using strain EG7765 that contains the genetic marker oxTi76 (oxTi76 contains a transposon insertion of Peft-3::GFP::H2B::tbb-2 3’UTR on chromosome VI, in a location close to the flp-1 gene) and animals of the F2 generation that were not green, and did not exhibit Daf-c or elevated body curvature were picked. This outcross resulted in the isolation of strain XZ2394 that contains the flp-1(ok2811) mutation but does not show the same extent of elevated body curvature as XZ2314 (flp-1(ok2811)) strain (Fig. 1G). This suggests that strain RB2126, besides the mutation in daf-11, contains an additional mutation that partially but significantly suppresses the flp-1(ok2811) elevated body posture. This new allele was named yak156 and the updated genotype of strain RB2126 is flp-1(ok2811); daf-11(yak155); yak156.
Methods
Request a detailed protocolStrain maintenance
Strains were maintained at 15˚C or room temperature (23˚C) on NGM plates with E. coli strain OP50 as a food source (Brenner, 1974).
Waveform quantification
First-day adult animals were placed on an OP50 plate and allowed to move forward until they had completed at least five tracks. Each animal’s tracks were imaged at 50X magnification using a Nikon SMZ745 stereoscope and an iLabCam. Worm track period and 2X amplitude were measured using the line tool in Image J. For each worm, three to five periods and amplitudes were measured. Each experiment was performed on 5-10 worms.
Statistics
P values were determined using GraphPad Prism 9.0 (GraphPad Software). Normally distributed data sets requiring multiple comparisons were analyzed by a one-way ANOVA followed by a Bonferroni test. Normally distributed pairwise data comparisons were analyzed by two-tailed unpaired t tests.
Strains
Bristol N2: wild type
EG7765: oxTi76[Peft-3::GFP::H2B::tbb-2 3’UTR, unc-18(+)] IV; unc-18(md299) X
JT195: daf-11(sa195) V
JT6709: unc-42(e270) V daf-11(sa195) V
RB2126: flp-1(ok2811) IV; daf-11(yak155) V; yak156
XZ2314: flp-1(ok2811) IV [3X outcrossed from RB2126]
ΧΖ2370: oxTi76[Peft-3::GFP::H2B::tbb-2 3’UTR, unc-18(+)] IV; daf-11(yak155) V
XZ2380: flp-1(ok2811) IV; daf-11(sa195) V
XZ2394: flp-1(ok2811) IV; yak156
XZ2406: flp-1(ok2811) IV; daf-11(yak155) V
Acknowledgments
I am grateful to Michael Ailion for supporting the completion of this project in his lab, for mapping the yak155 mutation, and for comments and edits on the manuscript. Special thanks to Michael Crawford for critical reading of the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by an NIH grant R01 NS109476 to Michael Ailion and Jihong Bai.
References
Funding
NIH grant R01 NS109476
Reviewed By
AnonymousHistory
Received: April 18, 2021Revision received: April 24, 2021
Accepted: May 4, 2021
Published: May 9, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Topalidou, I (2021). Background mutation in strain RB2126 affects the locomotion behavior of flp-1 mutants. microPublication Biology. 10.17912/micropub.biology.000393.Download: RIS BibTeX