Abstract
Skin infection with the fungus Drechmeria coniospora leads to a transcriptional response in the worm epidermis. This involves an increased expression of a group of antimicrobial peptide (AMP) genes including those in the nlp-29 and cnc-2 clusters. The major pathways leading to the expression of these AMP genes have been well characterized and converge on the STAT transcription factor STA-2. We reported previously that expression in the epidermis of a constitutively active (gain of function, gf) form of the Gα protein GPA-12 (GPA-12gf) recapitulates much of the response to infection. To reveal parallel pathways activated by infection, we focus here on an effector gene that is not induced by GPA-12gf. This gene, ifas-1, encodes a protein with a fascin domain, associated with actin binding. Its induction upon fungal infection does not require sta-2. A transcriptional reporter revealed induction in the epidermis of ifas-1 by infection and wounding. Thus, ifas-1 represents part of a previously unexplored aspect of the innate immune response to infection.
Description
The natural fungal pathogen Drechmeria coniospora pierces the worm’s cuticle and its hyphae grow throughout the organism. In the epidermis, this triggers a rapid increase in the expression of genes from the nlp (for neuro-peptide-like protein) and cnc (caenacin) families. These genes encode structurally-related antimicrobial peptides (AMPs). We have defined major signalling pathways required for the regulation of nlp‑29 gene expression. Two of them, one specific for infection and the second also activated by wounding, act upstream of a highly conserved p38 MAPK signalling cascade. The induction of cnc‑2 upon infection, on the other hand, is independent of PMK-1/p38 MAPK, but requires DBL-1/TGFß produced by certain neuronal cells, acting via a non-canonical TGFß pathway in epidermal cells. The STAT transcription factor-like protein, STA‑2, is essential for both the PMK-1/p38 MAPK, and DBL-1/TGFß immune signalling pathways, to govern the transcriptional response to fungal infection in the epidermis (reviewed in Kim and Ewbank, 2018; Martineau et al., 2021).
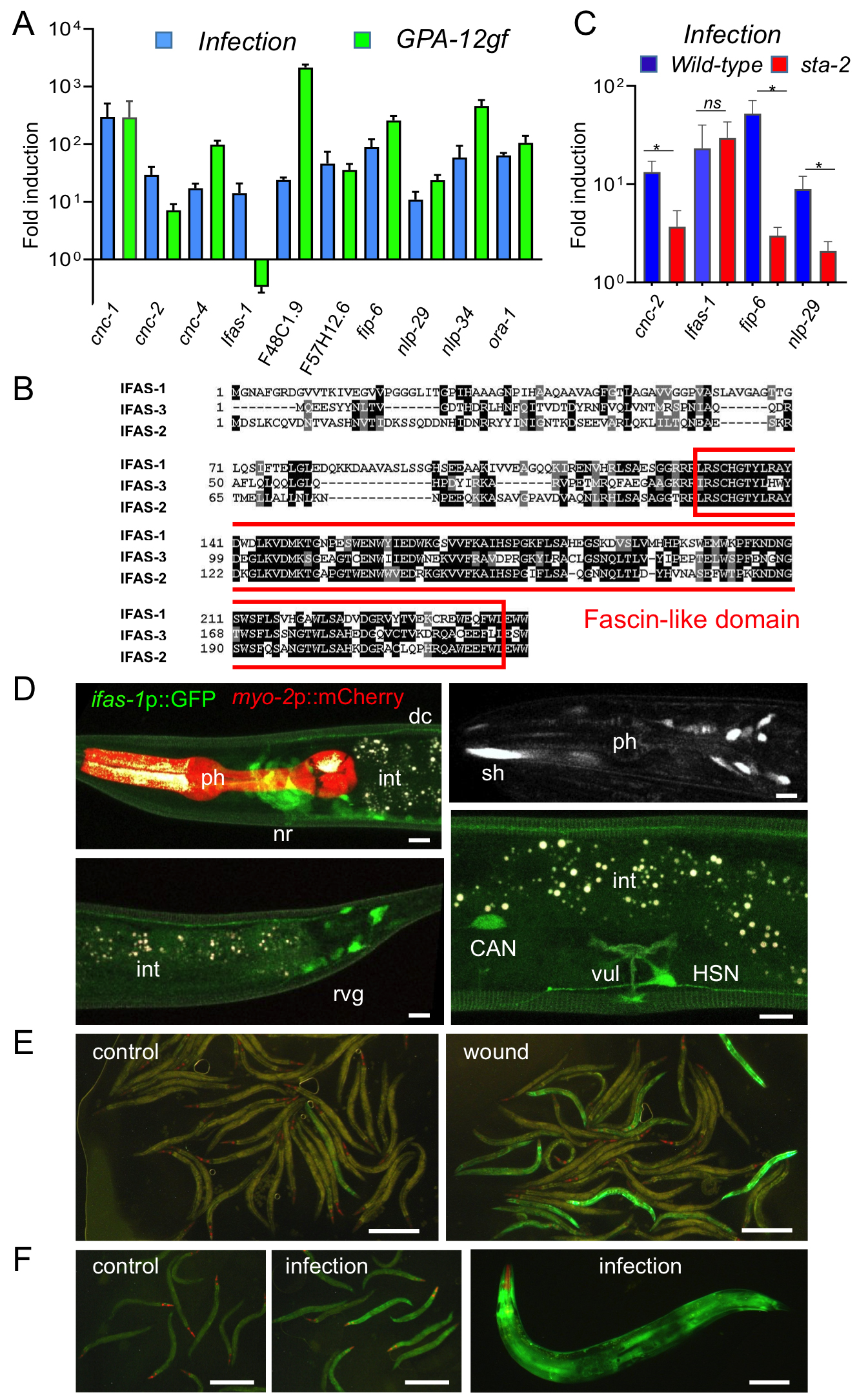
In the absence of infection, expression of a constitutively active Gα protein (GPA-12gf) in the adult epidermis leads to higher expression of AMP genes of both the nlp and cnc families (Labed et al., 2012). More generally, there is a considerable overlap between the genes induced by infection (Engelmann et al., 2011) and those up-regulated upon expression of GPA-12gf (Lee et al., 2018). To broaden our understanding of the host response to fungal infection, we selected 10 strongly-induced genes for validation through qRT-PCR. We confirmed that for 9 of them, expression was increased in both the infected and the GPA-12gf samples. They included members of the nlp and cnc families, but also several other genes predicted to encode small secreted peptides, like F48C1.9 (Omi and Pujol, 2019), fip-6 (Pujol et al., 2012), F57H12.6 and ora-1. For only one, F40H7.12, expression was induced by fungal infection but not in the GPA-12gf background (Figure 1A). The gene encodes a protein with a fascin domain, associated with actin binding. Two other C. elegans genes (F09C6.1 and Y105C5B.14) are predicted to encode proteins with a fascin domain (Figure 1B). Data in Wormbase indicates that their expression increases upon exposure to a variety of stresses. We therefore called this family ifas for “inducible fascin domain”. We determined that the induction of F40H7.12/ifas-1 upon fungal infection does not require sta-2, unlike nlp-29, cnc-2 or fip-6 (Figure 1C).
We made two different reporter transgenes to study the gene’s expression pattern, both containing the short (800 bp) intergenic region separating ifas-1 from its upstream gene, one with the 3’ UTR of unc-54, the other with its own 3’ UTR. Transgenic strains produced with the two constructs behaved similarly. A constitutive expression was observed in a subset of neurons including neurons in the head, lateral neurons, including the CAN neurons, the HSN neurons and a subset of retrovesicular ganglion neurons (Figure 1D). Upon fungal infection and wounding, an induction was observed in the epidermis. While the induction was robust and reproducible, it was only observed in less than 20% of the worms (Figure 1E-F). This may be because the reporter constructs do not contain all the regulatory elements required to reflect endogenous gene expression. Future characterization of ifas-1 is expected to reveal previously unexplored aspects of the innate immune response to epidermal fungal infection.
Methods
Request a detailed protocolMultiple Sequence Alignments of IFAS-1 (CE38709), IFAS-2 (CE15760) and IFAS-3 (CE24063) were done with MUSCLE https://www.ebi.ac.uk/Tools/msa/muscle/ and shaded with Boxshade https://embnet.vital-it.ch/software/BOX_form.html. Protein sequences correspond to those from WormBase release WS280.
Strains: All C. elegans strains were maintained on nematode growth medium (NGM) and fed with E. coli OP50, as described (Stiernagle, 2006): the wild-type N2, IG1570 frSi2[pNP138(col-19p::GPA-12gf), unc-119(+) ttTi5605] II; frIs7[nlp-29p::GFP, col-12p::DsRed] IV (Lee et al., 2018), IG1241 sta-2(ok1860) V (Dierking et al., 2011).
Constructs: pNP150 (ifas-1p::GFP::3’UTRunc-54) is derived from pCFJ151 that was a gift from Erik Jorgensen (Addgene plasmid # 19330; http://n2t.net/addgene:19330; RRID:Addgene_19330) (Frokjaer-Jensen et al., 2008). pNP150 (ifas-1p::GFP::3’UTRunc-54) was obtained by Gibson fusion of 800 bp of the ifas-1 promoter with primers cgactcactagtgggcagcctcaaaatactggatcac and gttcttctcctttactcatagcgttgcccatcagaaac. pNP153 (ifas-1p::GFP::3’UTRifas-1) was obtained by replacing the unc-54 3’UTR in pNP150 by 250 bp of the ifas-1 3’UTR with by Gibson fusion with primers ggatgaactatacaaatagtggtgatccatatttgtaag and gagaatgtctagaactaggcacccaacaaagttagctagc. Each construct was independently injected at 20 ng/µl together with pCFJ90 myo-2p::mCherry at 2 ng/µl, pZX13 at 20 ng/µl, pBSKS empty vector at 60 ng/µl. pCFJ90 was a gift from Erik Jorgensen (Addgene plasmid # 19327; http://n2t.net/addgene:19327; RRID:Addgene_19327) (Frokjaer-Jensen et al., 2008), pZX13 contains the hygromycin resistance gene HygR under the control of a minimal 380 bp rsp-0 promoter sequence (atttttgctttcgtcgtaaa to aatatgtcaggcggtgccgc). It was derived from SG120 (a kind gift of Jason Chin; Radman et al., 2013) by removing the B0393.2 gene, to decrease lethality associated with the original plasmid (S. O. unpublished observations). Two independent transgenic strains were obtained IG2065 frEx646[pNP150(ifas-1p::GFP::3’UTRunc-54), myo-2p::mCherry, rps-0p::HygR] and IG2066 frEx647[pNP153(ifas-1p::GFP::3’UTRifas-1), myo-2p::mCherry, rps-0p::HygR]. Images for IG2065 are presented in Figure 1 D-F.
Images were taken of worms mounted on a 2% agarose pad on a glass slide, anesthetized with 0.25 mM levamisole, using either a Leica MZ16 F stereomicroscope, a Zeiss LSM780 confocal microscope or a Visitron spinning disk, as previously described (Taffoni et al., 2020).
Infection & qRT-PCR: Worms were synchronised by the standard bleach method and exposed to fungal spores for 8 h at the L4 stage, or wounded with a microinjection needle at the young adult stage, as previously described (Pujol et al., 2008). RNA extraction and qRT-PCR were done with transcript specific primers, as previously described (Pujol et al., 2008); 3 replicates were analysed.
Reagents
qRT PCR primers:
| name | gene | sequence | WormBase associated Gene ID |
| 1087 | cnc-1 F | CTGCGCAATGGGGATATAACTCA | WBGene00000555 |
| 1088 | cnc-1 R | GAGAAGACCACCTCCACCAT | WBGene00000555 |
| 944 | cnc-2 F | CCGCTCAATATGGTTATGGAG | WBGene00000556 |
| 549 | cnc-2 R | TCCCATGCCCATACCGTAAC | WBGene00000556 |
| 1124 | cnc-4 F | ACAATGGGGCTACGGTCCATAT | WBGene00000558 |
| 1125 | cnc-4 R | ACTTTCCAATGAGCATTCCGAGGA | WBGene00000558 |
| 2340 | ifas-1 F | TTCCTGAGTGCTCACGAAGG | WBGene00044379 |
| 2341 | ifas-1 R | AACACTGAGGAACGACCAGG | WBGene00044379 |
| 2189 | F48C1.9 F | CCAATTAAGTACAGCTGCAA | WBGene00018601 |
| 2190 | F48C1.9 R | GTATCCAGGATAACTGTAATAG | WBGene00018601 |
| 2338 | F57H12.6 F | GGAAGAAGATCTCCACCTTG | WBGene00019021 |
| 2339 | F57H12.6 R | AATCGATAACTTCACGAGTC | WBGene00019021 |
| 2328 | fip-6 F | TGCAATTGTAACATACGCAC | WBGene00009964 |
| 2590 | fip-6 R | TAATATGGTTGATATCCACC | WBGene00009964 |
| 952 | nlp-29 F | TATGGAAGAGGATATGGAGGATATG | WBGene00003767 |
| 848 | nlp-29 R | TCCATGTATTTACTTTCCCCATCC | WBGene00003767 |
| 969 | nlp-34 F | ATATGGATACCGCCCGTACG | WBGene00015046 |
| 970 | nlp-34 R | CTATTTTCCCCATCCGTATCC | WBGene00015046 |
| 2336 | ora-1 F | CAAAGACAAGGAATCGAAGC | WBGene00003879 |
| 2337 | ora-1 R | TCATCCTTCACGTTCTCATC | WBGene00003879 |
Acknowledgments
We thank A. Bonnet, M. Bulle and S. Lee for technical assistance, E. Jorgensen & J. Chin for plasmids and J. Ewbank for constructive comments. We thank the imaging core facility (ImagImm) of the Centre d’Immunologie de Marseille-Luminy (CIML) supported by the French National Research Agency program (France-BioImaging ANR-10-INBS-04).
References
Funding
French National Research Agency (ANR-16-CE15-0001-01, ANR-16-CONV-0001) and institutional grants from CNRS, INSERM and Aix Marseille University to the CIML.
Reviewed By
AnonymousHistory
Received: April 2, 2021Revision received: May 18, 2021
Accepted: May 19, 2021
Published: May 25, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Omi, S; Zhang, X; Thakur, N; Pujol, N (2021). ifas-1 is upregulated by fungal infection in a GPA-12 and STA-2-independent manner in the Caenorhabditis elegans epidermis. microPublication Biology. 10.17912/micropub.biology.000400.Download: RIS BibTeX