Laboratory of Behavioral and Developmental Genetics, Department of Human Genetics, KULeuven, University of Leuven, B-3000 Leuven, Belgium
Department of Molecular Biology & Genetics, Cornell University, Ithaca NY 14853 USA
FlyBase, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3DY, U.K.
Abstract
The neprilysin (M13) family of metalloendopeptidases comprises highly conserved ectoenzymes that cleave and thereby inactivate many physiologically relevant peptides in the extracellular space. Impaired neprilysin activity is associated with numerous human diseases. Here, we present a comprehensive list and classification of M13 family members in Drosophila melanogaster. Seven Neprilysin (Nep) genes encode active peptidases, while 21 Neprilysin-like (Nepl) genes encode proteins predicted to be catalytically inactive. RNAseq data demonstrate that all 28 genes are expressed during development, often in a tissue-specific pattern. Most Nep proteins possess a transmembrane domain, whereas almost all Nepl proteins are predicted to be secreted.
Description
Neprilysins belong to the family of M13 metalloendopeptidases and constitute highly conserved ectoenzymes that cleave and thereby inactivate numerous physiologically relevant peptides in the extracellular space. While the vast majority of these enzymes appear to be membrane-bound, some family members have been identified as soluble secreted proteins (Turner et al., 2001). In humans, seven members of the M13 family are known, namely Neprilysin (NEP), endothelin-converting enzymes (ECE1, ECE2), ECEL1, MMEL1, the KELL blood-group protein and PHEX (Turner and Tanzawa 1997). Among these, NEP is characterized best, with identified substrates including endothelins, angiotensins I and II, enkephalins, bradykinin, atrial natriuretic peptide, substance P and the amyloid-beta peptide (Turner et al., 2001, Nalivaeva et al., 2020). NEP-mediated hydrolysis is critical to maintain the physiological homeostasis of these peptides, and thus represents a prerequisite for proper endocrine signal transmission. Accordingly, impaired neprilysin activity in humans is involved in the pathogenesis of numerous diseases, including hypertension (Molinaro et al., 2002), analgesia (Whitworth 2003), cancer (Turner et al., 2001) and Alzheimer’s disease (Iwata et al., 2000, Belyaev et al., 2009). Clinical trials have confirmed the therapeutic potential of modulating neprilysin activity (Jessup 2014).
Previous analyses of the Drosophila melanogaster (hereafter, ‘Drosophila’) genome have identified up to 24 genes encoding M13 family members (Coates et al., 2000, Isaac et al., 2000, Bland et al., 2008, Sitnik et al., 2014). Five of these (Nep1–Nep5) are thought to encode catalytically active neprilysins. However, this has so far been demonstrated only for Nep2 and Nep4 (Thomas et al., 2005, Bland et al., 2007, Meyer et al., 2009, Hallier et al., 2016), and in vivo substrates are known only for Nep4 (Hallier et al., 2016). Nep2 is involved in the regulation of locomotion and geotactic behavior (Bland et al., 2009) and is required for early embryonic development (Sitnik et al., 2014). Nep4 is implicated in sustaining muscle integrity (Panz et al., 2012) and controls insulin signaling and feeding behavior (Hallier et al., 2016). Neprilysin activity in general appears to be critical to the formation of middle- and long-term memory (Turrel et al., 2016), reproduction (Sitnik et al., 2014) and regulation of pigment dispersing factor signaling within circadian neural circuits (Isaac et al., 2007). Significantly, increased expression of Nep1 or Nep2 ameliorates the detrimental effects of amyloid-beta peptide overexpression in Drosophila models of Alzheimer’s disease (Finelli et al., 2004, Cao et al., 2008, Sofola-Adesakin et al., 2016, Turrel et al., 2017, Turrel et al., 2020). Other Drosophila M13 members lack key catalytic residues and are therefore predicted to be inactive or have non-enzymatic functions (Bland et al., 2008, Sitnik et al., 2014). These ‘neprilysin-like’ proteins include: Fra mauro (Frma), which is involved in sex peptide responses and is required for female remating receptivity and fertility (Findlay et al., 2014); Gone early (Goe), which functions in the ovary to limit the number of germline stem cells (Matsuoka et al., 2014); and CG4721, which plays a role in eye development and is also involved in lipid and carbohydrate storage (Nfonsam et al., 2012, Banerjee et al., 2021). While these studies have clearly advanced the current understanding of M13 family functionality in Drosophila, the overall physiological relevance of individual members is still far from being understood.
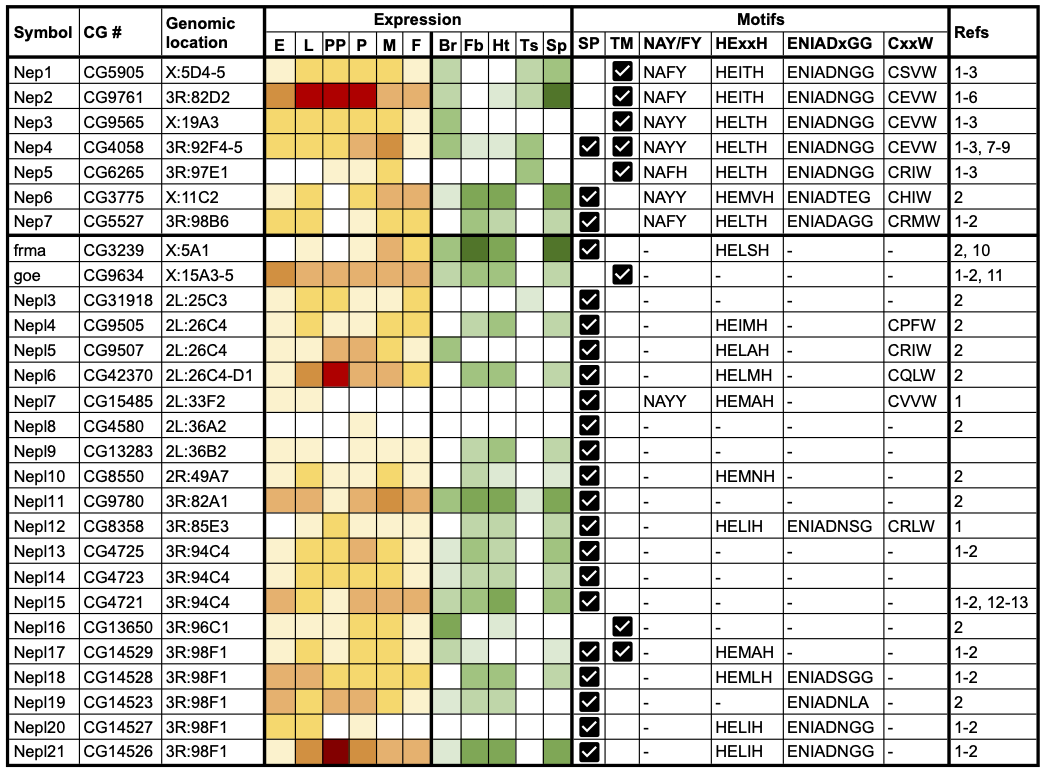
We aimed to generate a comprehensive and up-to-date list of Drosophila M13 family members and systematically assess evidence for their expression and functional activity. We applied a combination of literature review and bioinformatic analyses (see Methods) to identify a total of 28 Drosophila M13 genes, including two (CG13283 and CG4723) that had not been identified in previous studies (Table 1). These were classified into seven neprilysin (Nep) and 21 neprilysin-like (Nepl) genes and named accordingly. Nep classification required the presence of four conserved sequence motifs in the encoded proteins that are critical to catalytic activity in vertebrate neprilysins: HExxH and ENIAD(xGG) represent zinc-binding domains, CxxW is critical to protein folding and maturation, and NAY/FY mediates substrate or inhibitor binding (Turner et al., 2001, Sitnik et al., 2014). The Nep genes comprise the previously named Nep1, Nep2, Nep3, Nep4 and Nep5 genes, together with CG3775 (Nep6) and CG5527 (Nep7). The 21 Nepl-genes encode proteins that exhibit significant similarity to neprilysins in their primary structure, but lack one or more of the motifs required for catalysis. The symbols of frma (CG3239) and goe (CG9634) have not been changed, but ‘Nepl1’ and ‘Nepl2’ have been added as respective synonyms to recognize the fact they were the first and second Nepl genes to be characterized. The remaining Nepl genes have been given a numerical suffix, incremented based on their genomic location. The revised gene nomenclature has been incorporated into FlyBase (https://flybase.org, Larkin et al., 2021).
It is evident that Drosophila has an expanded set of Nep and Nepl genes compared to mammals (Coates et al., 2000, Bland et al., 2008). The 28 Drosophila genes are distributed throughout the genome and are present on all major chromosome scaffolds except chromosome arm 3L (Table 1). Three distinct clusters of Nepl genes are evident. Nepl4, Nepl5 and Nepl6 are located in a 17.6 kb interval (also containing two other genes) at cytological position 26C4-D1 on 2L; Nepl13, Nepl14 and Nepl15 are arranged as tandem repeats in head-to-tail orientation in a 7.4 kb interval at 94C4 on 3R; and Nepl17, Nepl18, Nepl19, Nepl20 and Nepl21 are arranged as tandem repeats in a 13.1 kb interval at 98F1 on 3R. These Nepl clusters are likely to be the result of local duplication events, consistent with previous phylogenetic analyses (Bland et al., 2008, Sitnik et al., 2014).
We analysed genome-wide RNAseq data (Graveley et al., 2011, Leader et al., 2018) to systematically examine evidence for Nep and Nepl gene expression. As summarized in Table 1, all 28 genes are expressed at some point during development (yellow/red heatmap) and exhibit tissue-specific expression in adults, notably within the brain/CNS, thoracicoabdominal ganglion, fat body, heart and reproductive tracts (green heatmap). For the Nep genes, Nep1-4 are expressed throughout development, with Nep2 being the most highly expressed during the larval-pupal period, whereas Nep5-7 are only detected at discrete stages. All Nep genes, except Nep3, are expressed in the male and/or female reproductive tract, with Nep2 having particularly high expression in spermathecae. Most Nep genes are also expressed in the brain/CNS, while around half show expression in the fat body and/or heart. A similar pattern is seen for the Nepl genes: most are expressed at all developmental stages, though expression of Nepl7 and Nepl8 is undetectable/extremely low throughout development. Nepl6 and Nepl21 are notable for their high expression in prepupae, suggesting an important role during metamorphosis. Most Nepl genes are expressed in the spermatheca, while only two (Nepl3 and Nepl11) are detectable in the testis. A major share is also expressed in the fat body and heart, and around half of the Nepl genes are expressed in the brain/CNS. Amongst the Nepl genes, frma is expressed at particularly high levels within the fat body and spermathecae. Overall, these expression data are consistent with previous reports (Thomas et al., 2005, Bland et al., 2007, Iijima-Ando et al., 2008, Meyer et al., 2009, Meyer et al., 2011, Findlay et al., 2014, Matsuoka et al., 2014, Sitnik et al., 2014, Banerjee et al., 2021), and further demonstrate that all 28 Nep and Nepl genes are actively transcribed and therefore potentially functional, likely acting in a stage- and/or tissue-specific manner.
Finally, we examined Nep/Nepl protein sequences for evidence of a signal peptide or transmembrane domain that would indicate the proteins are secreted or membrane-localized, respectively (see Methods). Remarkably, two Nep and the majority (18) of Nepl proteins lack a predicted transmembrane domain and, instead, possess a predicted signal peptide suggesting that they are secreted (Table 1). Although Nep2 is predicted to possess a transmembrane domain, experimental data indicate that it is secreted, presumably via proteolytic cleavage of a membrane-localized proprotein (Thomas et al., 2005). Nep4 is unique in encoding either a transmembrane or a secreted isoform as a result of alternative splicing (Meyer et al., 2009), while Nepl17 encodes a single isoform containing both a signal peptide and a transmembrane domain. One function of the secreted Nepl proteins may be to bind and sequester peptide targets of the catalytically active neprilysins. Such a biological role has already been suggested for Nepl15 (Banerjee et al., 2021) and could establish a novel facet of the neprilysin-mediated regulation of peptide homeostasis.
In summary, we have combined bioinformatic analyses with an evaluation of relevant publications to generate a comprehensive list and classification of genes encoding neprilysin and neprilysin-like proteins in Drosophila. The respective dataset has been compiled as a FlyBase gene group report (https://flybase.org/reports/FBgg0000963.html). These resources will support further research and understanding of the biological roles of Nep and Nepl proteins in flies. Identifying the physiological functions and substrates of this set of largely uncharacterized proteins in Drosophila may provide clinically relevant insights into neprilysin function in humans.
Methods
Request a detailed protocolPublications identifying/characterizing Drosophila neprilysins were identified using PubMed (https://pubmed.ncbi.nlm.nih.gov) and FlyBase (https://flybase.org, Larkin et al., 2021). Previously published lists of Drosophila neprilysins were obtained from Bland et al., 2008, Meyer et al., 2011 and Sitnik et al., 2014. De novo identification of Drosophila neprilysins was performed using three approaches: (i) searching FlyBase (FB2021_02) for Drosophila proteins containing the InterPro signature “Peptidase M13 family (IPR000718)” (Blum et al., 2021); (ii) identifying orthologs of human M13 peptidases (MME (aka NEP), MMEL1, ECE2, ECE1, KEL, PHEX, ECEL1) using the integrative ortholog prediction tool, DIOPT (v8.0) (Hu et al., 2011) via FlyBase; (iii) querying the MEROPS database (release 12.3; https://www.ebi.ac.uk/merops/; Rawlings et al., 2018) for D. melanogaster members of the M13 peptidase family. Gene symbol and genomic location data were obtained from FlyBase (FB2021_02). Developmental stage expression data are derived from the modENCODE developmental transcriptome RNAseq dataset (Graveley et al., 2011), as implemented as heatmaps within FlyBase; Table 1 shows the mean expression levels for each major developmental stage. Adult tissue expression data and heatmaps are from FlyAtlas2 (http://flyatlas.gla.ac.uk/FlyAtlas2/index.html; Leader et al., 2018). Male and female tissue-specific data were quantitatively and qualitatively similar, as were data for the brain/CNS and thoracicoabdominal ganglion, and virgin and mated spermathecae; thus, only representative data for female tissues, brain/CNS and mated spermathecae are shown in Table 1. Relatively little Nep and Nepl expression is seen in the accessory gland or ovary, and so expression in those tissues is not reported in Table 1. Sequence analysis and motif identification was done using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), SignalP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP-5.0/) and Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan) with all available motif databases being selected. In addition, all sequences were manually analyzed for the presence of respective motifs, combined with sequence alignments (http://multalin.toulouse.inra.fr/multalin/) to ensure proper motif localization.
Acknowledgments
We thank Achim Paululat and Maik Drechsler for valuable comments on the manuscript.
References
Funding
This research was funded by the German Research Foundation (SFB 944: Physiology and Dynamics of Cellular Microcompartments) to HM (P21); grants from the National Institute of Child Health and Development of the NIH [R37HD038921 to MFW (PI) and R01HD059060 to MFW and Andrew Clark (PIs)]; FWO grants G065408.N10 and G078914N and a KULeuven grant C14/17/099 to PC; and a stipend from the Hans Mühlenhoff Foundation to RS. SJM is funded by a grant from the National Human Genome Research Institute of the NIH [U41HG000739] to Norbert Perrimon (PI), Nicholas Brown (co-PI).
Reviewed By
AnonymousHistory
Received: June 4, 2021Revision received: June 16, 2021
Accepted: June 16, 2021
Published: June 23, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Meyer, H; Buhr, A; Callaerts, P; Schiemann, R; Wolfner, MF; Marygold, SJ (2021). Identification and bioinformatic analysis of neprilysin and neprilysin-like metalloendopeptidases in Drosophila melanogaster. microPublication Biology. 10.17912/micropub.biology.000410.Download: RIS BibTeX