Abstract
Drosophila larval crawling is easily-observable and relatively stereotyped. Crawling consists of linear locomotion interrupted by periods when the larvae pause, swing their heads, and change direction (a ‘search’). Here we identify Numb, a peripheral membrane adaptor protein, as an important regulator of searching behavior. When Numb RNAi transgenes were expressed in all neurons, searching frequency increased while linear movement appeared normal. Numb’s role in suppressing searching behavior was verified by rescuing this phenotype with a Numb homologue from mice. Such behavioral specificity suggests that further analysis of searching might help identify additional, evolutionarily-conserved interactors of the Numb protein.
Description
Numb was first identified as an adaptor protein required for Drosophila melanogaster sensory organ formation (Uemura et al., 1989). Over time, Numb and its binding partners have been implicated in a variety of cellular processes, including neurogenesis (Zhong et al., 1996), chemotaxis (P. Zhou et al., 2011), axon outgrowth (Huang et al., 2005), synapse formation (L. Zhou et al., 2015), social behavior (Wang et al., 2019), and cancer (Pece et al., 2011).
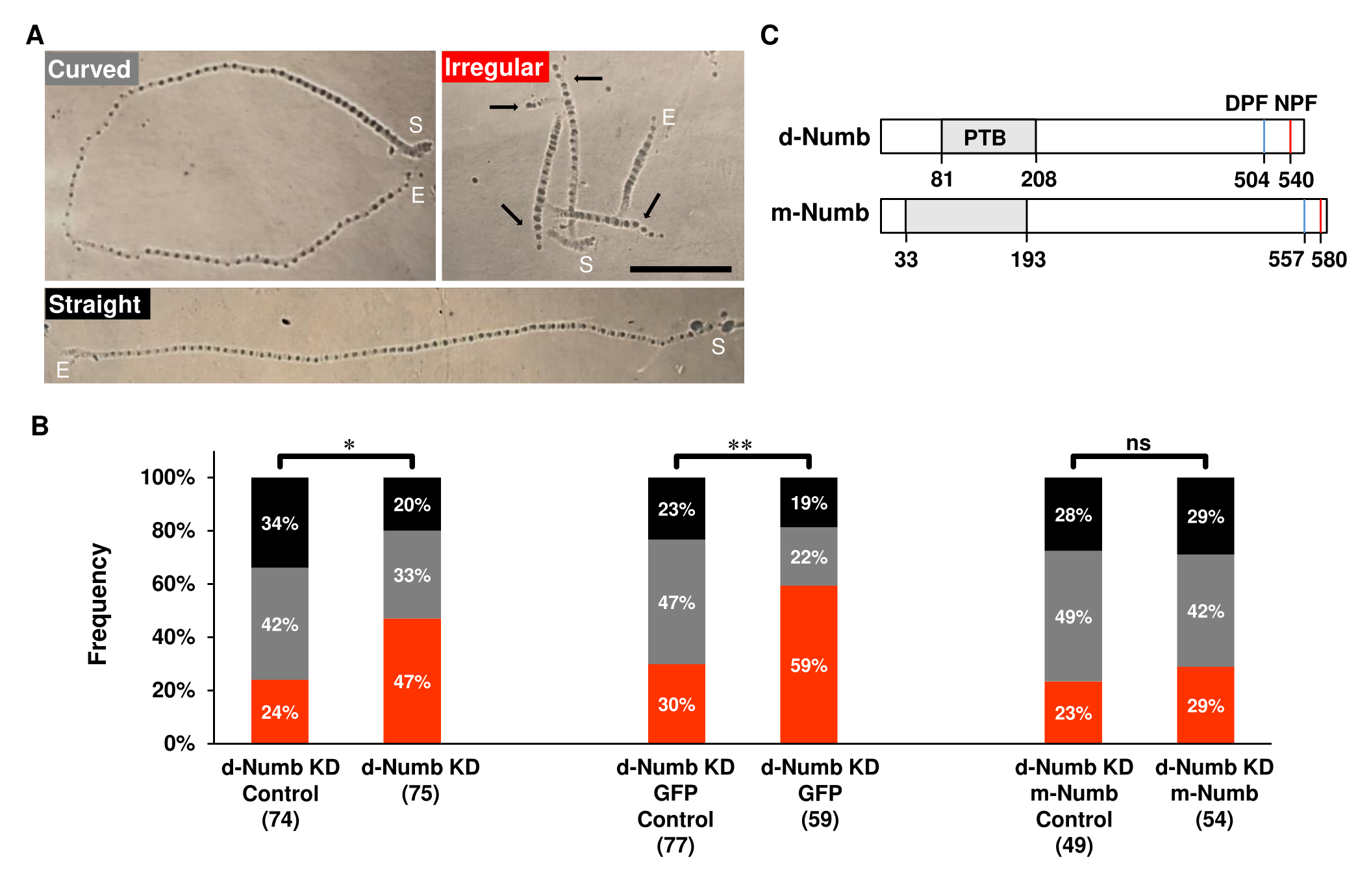
In Drosophila, loss of function Numb mutations alter the development of both central and peripheral neurons, including the networks that mediate larval crawling behavior (Spana et al., 1995, 1995; Tang et al., 2005; Vervoort et al., 1997). Larvae crawl through peristaltic waves of posterior to anterior body wall contractions that hydrostatically push the head forward to allow the animal’s mouth hooks to anchor at a new location on the substrate. Forward (and backward) crawling requires the activity of central pattern generators within the larval ventral ganglia (Clark et al., 2018; Fox et al., 2006). These networks set the rate and intensity of body wall contractions (Wang et al., 1997, 2002), integrating sensory feedback from multidendritic cells and chordotonal organs in the larval body wall (Hughes & Thomas, 2007; Ohyama et al., 2013; Song et al., 2007). Periodically, larvae will pause, swing their head, and begin crawling in a new direction (called here, a ‘search’). The mixture of linear crawling and searching behavior leads to several distinct crawling trajectories that can be identified by video (Heckscher et al., 2012; Wang et al., 2002) and by the pattern of mouth hook imprints when larvae crawl on soft substrates (Fig. 1A). Larval crawling has been extensively studied with genetic tools. However, most mutational and neural circuit dissection techniques have uncovered the biology of linear crawling, with fewer genes and cells being implicated in searching behavior.
Here, we show that Numb regulates the searching behavior of wandering-stage larvae. Numb’s role in neuronal development causes null mutations to be lethal, so we knocked down Drosophila Numb (d-Numb KD) selectively in neurons using RNAi transgenes (UAS-Numbds 5,10; Tang et al., 2005). Numb RNAi was expressed when the inducer, RU-486, was added to the culture media (Berke et al., 2013; Nicholson et al., 2008; Osterwalder et al., 2001; Roman et al., 2001). The crawling behavior of these experimental animals was compared to that of control larvae reared on the same food but without RU-486. Wandering-stage larvae were identified on the sides of culture vials, washed, and gently placed on 0.5% agarose plates to crawl for 2min. Videos and photos of the crawling trajectory and searching behavior (indicated in Fig. 1A) were compared (Methods). Total distance traveled, and hence average velocity, was assessed in straight and curved paths that lacked searches. NumbKD did not alter the distance crawled [Mean±SEM(n); control: 7.52±0.64cm(29) vs Numb KD: 7.56±0.40cm(21)]. However, NumbKD significantly increased larval searching. In both groups, the number of searches per 2min trial ranged from 0-9, but 31 of 75 (41%) of Numb KD animals searched 3 or more times while only 18 of 74 (24%) of the control showed this level of searching behavior (p=0.0046). The bias towards increased searching resulted in a significantly higher proportion of irregular path types, characterized by sharp turns and short linear segments between turns (Fig. 1B, left hand pair of bars, p = 0.016).
To verify that the increase in searching behavior was due to a reduction in Drosophila Numb (d-Numb) expression, we attempted to rescue this defect by co-expressing an RNAi-insensitive homolog from mouse (m-Numb; Numb4; UAS-numb4; Tang et al., 2005). Like d-Numb, the mouse isoforms contain the conserved DPF and NPF motifs, involved in membrane recycling (Santolini et al., 2000), and they share significant homology within the PTB domain, known to recruit E3 ligases (Dho et al., 1998; Fig 1C). m-Numb also contains a putative proline rich region (PRR) (aa 183-555; not shown in Fig. 1), which interacts with SH3 domains of partner proteins (Luo et al., 2020). The mammalian isoform used for rescue of the searching phenotype (m-Numb4; Huang et al., 2005) has been used previously to rescue defects in cellular differentiation and fate selection in vivo (Tang et al., 2005; Verdi et al., 1999; Zhong et al., 1996). We first verified that the addition of a second UAS transgene (UAS-mCD8-GFP) would not diminish the increased search behavior caused by NumbKD (Fig. 1B, middle pair of bars). When m-Numb replaced GFP as the second downstream UAS transgene, the hyperactive searching behavior was significantly reduced to control levels (trajectories with 3+ searches; p=0.2307) leading to a distribution of path types similar to that seen with the control (Fig. 1B, right hand pair of bars; p = 0.90). This rescue is in line with the conserved function of Numb between flies and mice (Zhong et al., 1996).
The regulation of searching behavior in Drosophila larvae is both complex and incompletely understood. Sensory feedback from peripheral chordotonal organs and multi-dendritic neurons can initiate searching (Ainsley et al., 2003; Guo et al., 2016; Ohyama et al., 2013), the behavior can be stimulated by a small population of ventral ganglia interneurons (Clark et al., 2016), and searching is influenced by several neurotransmitter systems (Okusawa et al., 2014; Saraswati et al., 2004; Selcho et al., 2012; Suster et al., 2004). Numb may influence one or more of these regulatory mechanisms, and more specific RNAi expression would be instructive. The inducible nature of the GeneSwitch system and the ability to rescue the searching phenotype with m-Numb will provide interesting opportunities to dissect both the relevant spatial (cellular) and developmental aspects of Numb signaling. In this regard, it is possible that Numb-dependent protein trafficking, proteosomal-dependent degradation, endocytosis, axonal/synaptic targeting, or cellular differentiation may individually or coordinately cause this behavioral phenotype. Lastly, double-mutant analysis of larval searching may be an efficient assay to identify Numb’s evolutionarily-conserved interactors.
Methods
Request a detailed protocolFly stocks. All fly strains were cultured on a standard dextrose/cornmeal medium at 21ºC. ELAV-GS-GAL4 (FlyBase ID FBti0116249, a gift from H. Keshishian, Yale University) was used to express UAS constructs in all neurons. GeneSwitch (GS) is a bi-partite expression system that uses a conditional RU-486 dependent GAL4 protein to induce transgene expression (Berke et al., 2013; Osterwalder et al., 2001). The parents/progeny of experimentals and controls developed with and without 5µg/ml RU-486 in the media, respectively. UAS-Numbds5,10 and UAS-mNumb#4 (both gifts from W. Zhong, Yale University) were used to knockdown d-Numb and express m-Numb. UAS-CD8-GFP (FlyBase ID FBti0131931; Bloomington Stock Center, NIH P40OD018537) was used during the co-expression control experiments.
Larval locomotion. Wandering, third instar larvae were each gently removed from their culture vial, washed and sexed, and then placed in the center of 0.5% agarose plates at the start of each trial. The 2min trials began when productive crawling was observed and they were recorded using a Sony HDR-CX405 Camcorder (Sony Corporation, New York, NY). Only trials lasting the full time were used. Any trial in which larvae collided with the edge of the 14cm diameter plate were ended and not used. At the end of each trial, the animal was removed and the mouth hook imprints were photographed and used to categorize the type of trajectory. Locomotion was classified as ‘straight’ if the mouth hook imprints did not deviate from a straight line by more than 1cm from a line connecting the path’s start and end points. If so, the path was classified as ‘curved’. Any path that involved three or more searching events was classified as ‘irregular’. A ‘search’ was defined as the presence of a partially followed path (3 mouth hook imprints that were distinct from the main path) that was separated from other searches by at least three imprints. Path lengths for straight and curved paths with no searches were measured manually. Larvae were never re-tested during additional trials and animals were only taken from healthy culture vials. Over the course of the experiments, third instar larvae were taken from multiple culture vials and were the progeny of several sets of parental animals.
Statistics. All comparisons were examined with chi-square tests in Minitab 18 (Minitab, LLC., State College, PA). Statistical significance is indicated as p<0.05 (*), p<0.005 (**) and not significant (ns) in Fig. 1.
Acknowledgments
We thank Weimin Zhong and Haig Keshishian, both from Yale University, for providing fly stocks. We also acknowledge members of the Berke Lab and the Spring 2021 Reviewing Scientific Literature (BIO444) class at Truman State University for critical comments and suggestions.
References
Funding
This work was supported by a Grant In Aid of Student Research from Truman State University to AG, KR, NP, JB, and BB as well as laboratory start-up funds allotted by the Biology Department at Truman State University to BB.
Reviewed By
AnonymousHistory
Received: July 2, 2021Revision received: July 14, 2021
Accepted: July 15, 2021
Published: July 26, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Galbraith, A; Leone, S; Stuart, K; Emery, J; Renkemeyer, MK; Pritchett, N; Galbraith, L; Stuckmeyer, H; Berke, B (2021). Reducing the expression of the Numb adaptor protein in neurons increases the searching behavior of Drosophila larvae. microPublication Biology. 10.17912/micropub.biology.000426.Download: RIS BibTeX