Abstract
Heparan sulfate proteoglycans (HSPGs) are diverse macromolecules consisting of a protein core modified with glycosaminoglycan (GAG) chains. HSPGs, including glypicans and perlecans, have been implicated in shaping the extracellular matrix (ECM) to affect growth factor signaling. Here, we tested if GPN-1/glypican or UNC-52/perlecan plays a role in the bone morphogenetic protein (BMP) signaling pathway in patterning the C. elegans postembryonic mesoderm. Using the suppression of sma-9(0) (Susm) assay, we found that animals carrying mutant alleles of gpn-1 or unc-52 do not exhibit any Susm phenotype. We also tested and found that the two glypicans GPN-1 and LON-2 do not share functional redundancy in the BMP pathway. Our results suggest that GPN-1/glypican and UNC-52/perlecan do not play a major role in the C. elegans BMP pathway, at least in patterning of the postembryonic mesoderm.
Description
Heparan sulfate proteoglycans (HSPGs) are macromolecules composed of a protein core decorated with glycosaminoglycan (GAG) chains. These highly diverse molecules are classified based on their localization at the membrane or being secreted as part of the extracellular matrix (ECM), such as glypicans or perlecans, respectively (Sarrazin et al. 2011). HSPGs have been shown to play a structural role in the ECM as well as in the distribution of growth factors, such as transforming growth factor-β (TGF-β), fibroblast growth factor (FGF), Wnt, and Hedgehog (Hh), within tissues through highly specific binding interactions (Lin and Perrimon 2000). In vitro studies have demonstrated that human perlecan can bind to the BMP2 ligand (Decarlo et al. 2012). Understanding the biological roles of HSPGs in cell signaling has implications in disease diagnosis and potential treatment.
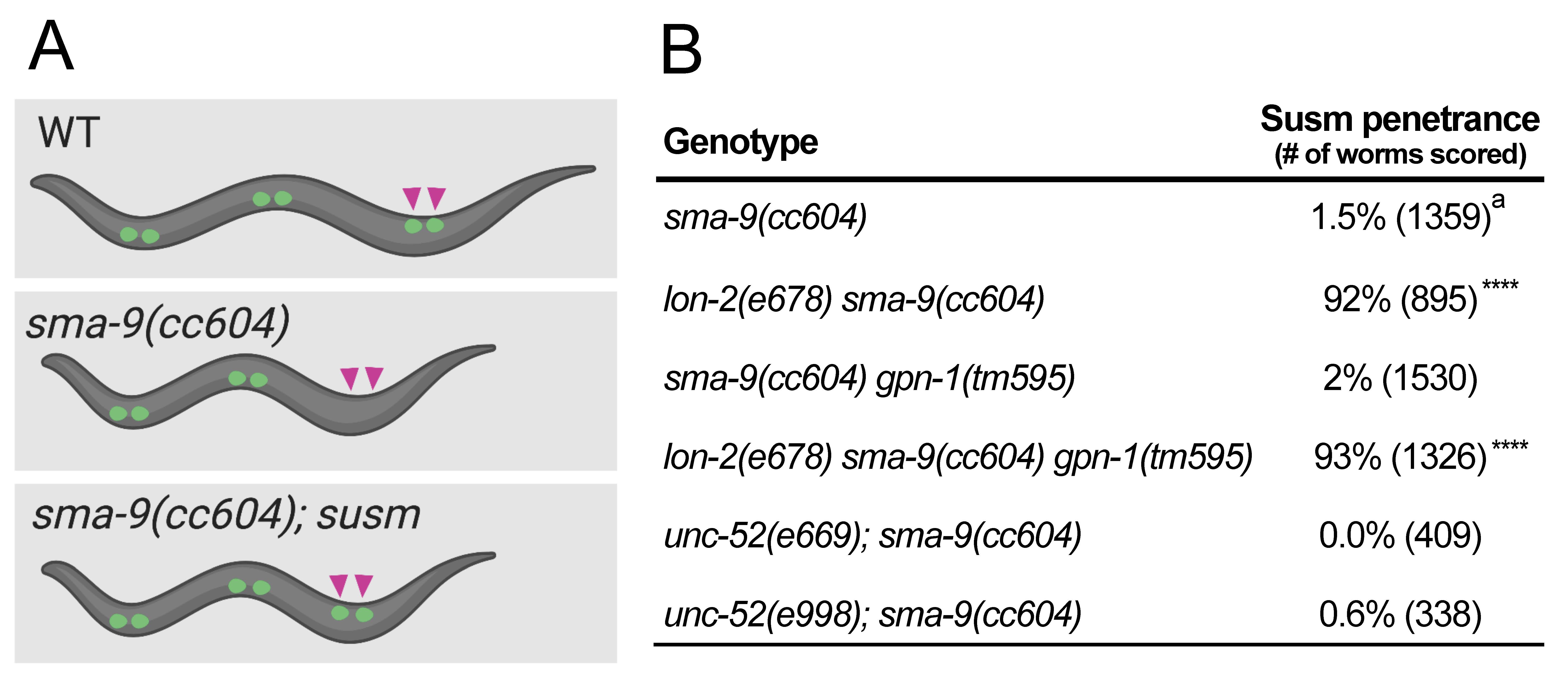
Bone morphogenetic protein (BMP) belongs to the TGF-β superfamily of signaling molecules. In C. elegans, the BMP signaling pathway is known to regulate multiple processes, including body size and postembryonic mesoderm patterning (Savage-Dunn and Padgett 2017). We have shown that mutations in the BMP pathway exhibit the Susm phenotype, namely, they can specifically suppress the mesoderm defects of sma-9(0) mutants (Figure 1A, Foehr et al. 2006). We further showed that the Susm assay is a highly specific and sensitive assay for BMP signaling defects (Liu et al. 2015). Previous work has demonstrated that LON-2/glypican negatively regulates the BMP pathway, and it has been postulated that LON-2 functions by binding to and sequestering the ligand (Gumienny et al. 2007). We have found that the lon-2(e678) null mutants exhibit the Susm phenotype (Figure 1B; Foehr et al. 2006; Liu et al. 2015). Here we aim to determine if GPN-1/glypican or UNC-52/perlecan plays a role in BMP signaling by using the Susm assay.
gpn-1 encodes a glypican, a membrane localized HSPG. gpn-1 is expressed in the pharynx and ventral nerve cord (VNC) during embryogenesis (Hudson et al. 2006). Previous studies have shown that gpn-1 mutants do not have a body size phenotype, and gpn-1 cannot substitute for lon-2, which encodes another glypican, to regulate body size (Gumienny et al. 2007; Taneja-Bageshwar et al. 2013). However, the Drosophila glypicans Dally and Dally-like are known to function in the BMP pathway by regulating the distribution of the BMP ligand within the ECM (Belenkaya et al. 2004; Norman et al. 2016). Furthermore, the C. elegans BMP ligand, DBL-1, is secreted from cells of the VNC (Suzuki et al. 1999), the same tissue that expresses gpn-1. We sought to test if gpn-1 plays a role in the BMP pathway by using the more sensitive Susm assay. We assayed for the Susm phenotype of gpn-1(tm595), a deletion allele lacking part of exon 2 and all of exon 3 of the gpn-1 gene. We found that gpn-1(tm595) animals exhibited a 2% penetrance of the Susm phenotype, within the range of background observed in the sma-9(cc604) single mutants (Foehr et al. 2006; Liu et al. 2015).
Previous studies have shown that gpn-1 and lon-2 act redundantly in hermaphrodite specific neuron migration (Kinnunen 2014). To test if these two glypicans may function redundantly in the BMP pathway, we examined the Susm phenotypes of lon-2(e678) gpn-1(tm595) double mutants. We found that gpn-1(tm595) does not enhance the penetrance of the Susm phenotype of lon-2(e678). Taken together, these results suggest that gpn-1 does not play a major role in the BMP pathway.
Perlecan is a secreted HSPG, characterized by its localization to the basement membrane. Human perlecan has been shown to be a key component of basement membranes that can bind and sequester growth factors, such as FGFs and VEGFs (Costell et al. 1999; Mongiat et al. 2000; Ishijima et al. 2012). While a study has demonstrated that TGF-β signaling regulates the expression of perlecan (Dodge et al. 1995), a role for perlecan in the TGF-β signaling pathway has not been shown. C. elegans has one homolog of the human perlecan: unc-52 (Rogalski et al. 1993). Localized to the basement membrane of body wall muscles (BWM), UNC-52 has been shown to play a critical role in the assembly and maintenance of the BWM myofilaments (Mullen et al. 1999). Interestingly, previous studies have shown that unc-52 genetically interacts with dbl-1/BMP during distal tip cell (DTC) migration (Merz et al. 2003). We tested if unc-52 plays a role in the BMP pathway by using the sma-9 suppression assay. We used two nonsense alleles of unc-52, e669 and e998, which introduce an early stop in exon 17 and 18, respectively. Both e669 and e998 were among the multiple unc-52 alleles used in the study by Merz et al. (2003). Neither unc-52(e669) nor unc-52(e998) exhibited any Susm phenotype (Figure 1B). Although not all UNC-52 isoforms are abolished in these two mutants, our results suggest that UNC-52/perlecan is unlikely to play a major role in the BMP pathway in C. elegans.
In summary, using the highly specific and sensitive Susm assay, we have found that unlike LON-2/glypican, GPN-1/glypican and UNC-52/perlecan do not appear to play a major role in the C. elegans BMP pathway. The C. elegans genome contains three other HSPGs, SDN-1/syndecan, AGR-1/agrin, and CLE-1/collagen XVIII (cle-1). It remains to be determined whether any of them play a role in BMP signaling.
Reagents
Strains:
| LW0040: | arIs37[secreted CC::gfp] I; cup-5(ar465) III; sma-9(cc604) X |
| LW5711: | arIs37[secreted CC::gfp] I; unc-52(e669) II; cup-5(ar465) III; sma-9(cc604) X isolate 1 |
| LW5712: | arIs37[secreted CC::gfp] I; unc-52(e669) II; cup-5(ar465) III; sma-9(cc604) X isolate 2 |
| LW5734: | arIs37[secreted CC::gfp] I; unc-52(e998) II; cup-5(ar465) III; sma-9(cc604) X isolate 1 |
| LW5735: | arIs37[secreted CC::gfp] I; unc-52(e998) II; cup-5(ar465) III; sma-9(cc604) X isolate 2 |
| LW5778: | arIs37[secreted CC::gfp] I; cup-5(ar465) III; lon-2(e678) sma-9(cc604) X isolate 1 |
| LW5779: | arIs37[secreted CC::gfp] I; cup-5(ar465) III; lon-2(e678) sma-9(cc604) X isolate 2 |
| LW5709: | arIs37[secreted CC::gfp] I; cup-5(ar465) III; sma-9(cc604) gpn-1(tm595) X isolate 1 |
| LW5710: | arIs37[secreted CC::gfp] I; cup-5(ar465) III; sma-9(cc604) gpn-1(tm595) X isolate 2 |
| LW5713: | ccIs4438 [intrinsic CC:::gfp] III; ayIs2[egl-15p::gfp] IV; lon-2(e678) sma-9(cc604) gpn-1(tm595) X isolate 1 |
| LW5714: | ccIs4438 [intrinsic CC:::gfp] III; lon-2(e678) sma-9(cc604) gpn-1(tm595) X isolate 2 |
Primers used for genotyping:
cc604: MLF-69: CGCAACAAGTTCATTCTCCA. MLF-70: CTTGGCTAAGATCCCATGCT. Sequence using LW-40: TCCGACTTGACACTTCATCAGC.
e678: JKL-1053: TTGTATTGCTCTACCGGTCC. JKL-1054: TTGCCCGGAATTTCAACTGC. JKL-1055: TCAACTTACGGAAGCGATCG.
e669: JKL-1871: TCCGTCAACTCTCTCGAAGG. JKL-1872: CACAAGTCGAAGCTCGTTAG. Sequence using JKL-1872.
e998: JKL-1873: CAAAATCGTTAGTGGCCGAG. JKL-1874: CCTGTTCACTCCCACTTCTC. Sequence using JKL-1873.
tm595: MSD-72: TGGCTTCACTGATTAGTACCGG. JKL-1868: ACCGTTCACATGGATCTTGAC. JKL-1870: CCATGCATACTCGCTGATCG.
References
Funding
This work was supported by the National Institutes of Health R35 GM130351 grant to J.L.. M.S.D. was partially supported by a National Science Foundation (NSF) Graduate Research Fellowship (DGE-1650441). R.G. was partially supported by a NYSTEM-RET Grant for Pre-College teachers.
Reviewed By
Cathy Savage-DunnHistory
Received: July 27, 2021Revision received: August 5, 2021
Accepted: August 10, 2021
Published: August 13, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
DeGroot, MS; Greer, R; Liu, J (2021). GPN-1/glypican and UNC-52/perlecan do not appear to function in BMP signaling to pattern the C. elegans postembryonic mesoderm. microPublication Biology. 10.17912/micropub.biology.000437.Download: RIS BibTeX