ETH Zürich
Abstract
The gpa-4 promoter-driven expression is described as specific for ASIL and ASIR chemosensory neurons in the nematode Caenorhabditis elegans, yet this was mostly examined in adult animals. Here we used a recombination-mediated reporter to test the previously used gpa-4 promoter expression. This reporter highlights all cells in which the gpa-4 promoter has been active at one point or another during development. We show that the gpa-4 promoter is indeed active in ASI, yet to our surprise, this promoter drives also expression in many other cell types, including the somatic gonad, the seam cells, a subset of tail and head neurons, and muscle cells, demonstrating a widespread activity of this transgenic gpa-4 promoter during embryonic and post-embryonic development.
Description
In C. elegans, the gpa-4 gene encodes a G-protein α-subunit, part of an evolutionary conserved Gα family expressed in muscle and neuronal cells (Jansen et al. 1999). Previous studies using fluorescent reporters have suggested that the gpa-4 promoter drives expression exclusively in the head chemosensory neurons ASIL and ASIR (Jansen et al. 1999). This led subsequent studies to consider the gpa-4 promoter as ASI-specific (see Table 1 for all studies using the gpa-4 promoter to drive gene expression).
Recently, red-to-green fluorescent reporter switches have been developed to validate promoter-driven transcriptional patterns and tissue specificity. These systems use either Cre/lox or FLP/FRT recombination. Their mode-of-operation, in principle, is identical and relies on two different transgenes. First, the recombinase is placed under transcriptional control of the promoter to be tested. Second, a ubiquitous promoter drives the expression of a red fluorophore, which is excised by the recombinase leading to the expression of a green fluorescent protein (Ruijtenberg and van den Heuvel 2015; Muñoz-Jiménez et al. 2017). As the red-to-green switch is irreversible, any cell or lineage in which the promoter has been active during development will be labeled green, while the rest of the animal will be red.
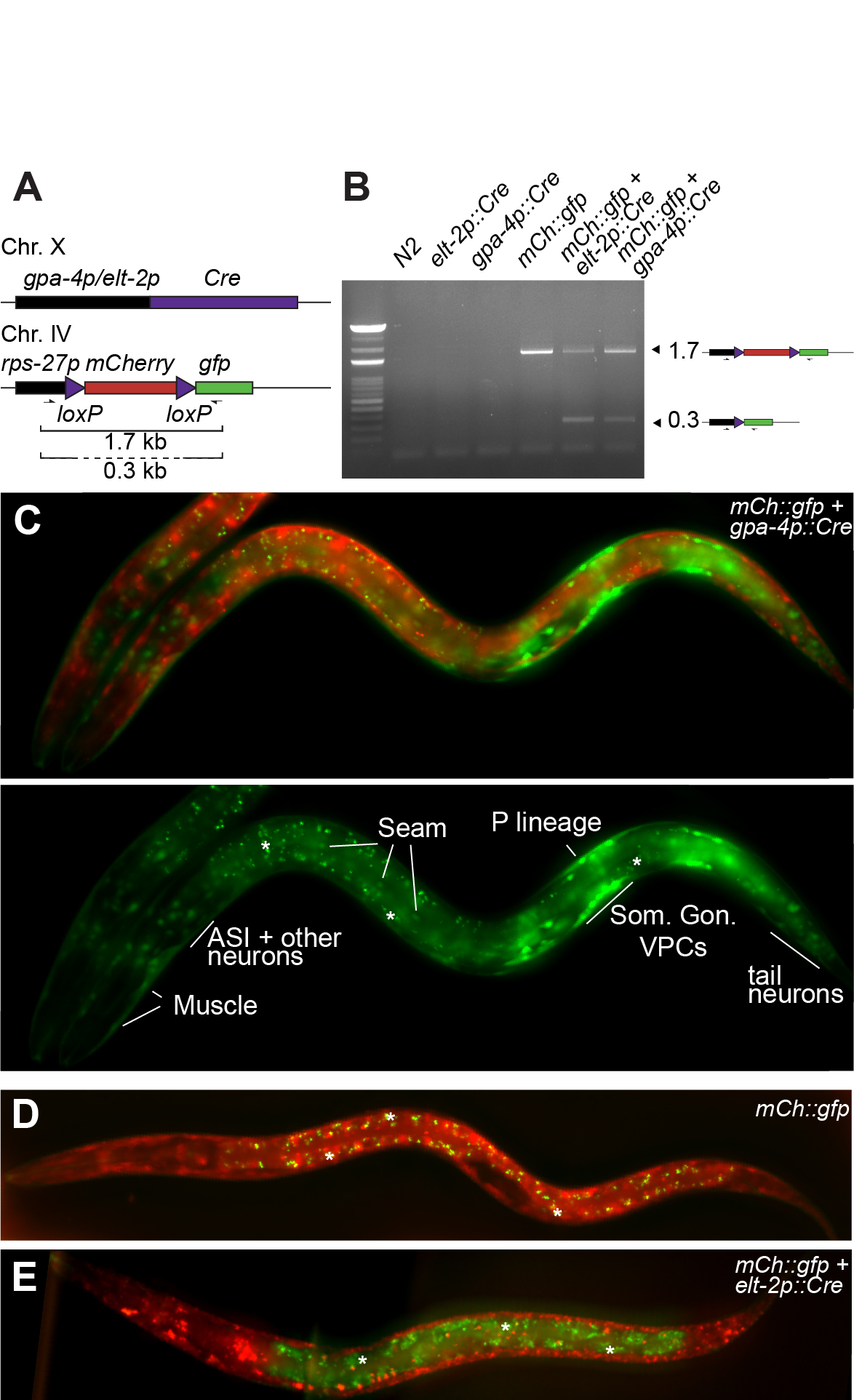
To examine the expression pattern of the gpa-4 promoter, we generated a gpa-4p::Cre transgene integrated on Chr X using MosSCI (Frøkjær-Jensen et al. 2012; Figure 1A). Recombination efficiency was first tested by PCR with primers encompassing the floxed mCherry cassette (Figure 1A,B). In the absence of recombination (mCherry::gfp cassette), a single product of 1.7 kb was amplified. When recombination occurs, two products will be visible, as most cells in the animal have the intact mCherry::gfp cassette while in the target tissue of the Cre driver construct, the mCherry cassette is deleted. This is indeed the case for both the positive control intestinal Cre driver (elt-2p::Cre) and the gpa-4p::Cre one, demonstrating that Cre is expressed and active when expressed under transcriptional control of the gpa-4 promoter. The intensity of the lower, recombined band was, however, surprisingly intense for an event which should only happen in the two ASI neurons per animal. Indeed, microscopic examination of animals showed clear and reproducible expression of GFP in many tissues, such as the somatic gonad, vulval precursor cells, seam cells, body wall muscle cells, tail and head neurons, as well as, the P lineage (Figure 1C). Both negative and positive controls showed the expected fluorescence pattern (no GFP and GFP expression only in the intestine, respectively; Figure 1D,E). Recombination by Cre driven by the gpa-4 promoter occurs therefore in many more cells than the published gpa-4 transcriptional fusion (Jansen et al. 1999) in which ASI-specific expression was reported.
One limitation of our approach could be that the insertion site of the gpa-4p::Cre influences its expression. However, there are two reasons arguing against this possibility. First, the same single copy insertion site was used for a variety of Cre drivers (muscle, intestine, mesoblast, vulva and XXX neuroendocrine cells; Ruijtenberg and van den Heuvel 2015; Gómez-Saldivar et al. 2020). Expression patterns for the different promoters did in each case correspond to the expected expression pattern. Second, single cell RNA-seq in L1 larvae (Cao et al. 2017) detected the endogenous gpa-4 transcript in cells identified as somatic gonad precursors, vulval precursors, sex myoblasts and ciliated sensory neurons (Extended data). These tissues overlap with the cell types in which we identified gpa-4 promoter activity and Cre expression. Taken together, we conclude that the previously thought ASI-specific transgenic gpa-4 promoter is widely active during embryonic and larval development.
Methods
Request a detailed protocolWorm growth
Worms were grown on solid nematode growth media (NGM), seeded with OP50 bacteria for maintenance culture and genetic crosses. All animals were grown at 22°C. Male animals were induced at 30ºC for 5 hours to carry out genetic crosses. For microscopy experiments using the iMIC platform, synchronized L1s were seeded onto 45 mm plates. Microscopy experiments were carried out after 12 hours.
Fluorescence imaging
Animals were laid on a pad of 2% agarose in water supplemented with 0.01% sodium azide, on a microscope slide, covered with a drop of M9 buffer and a coverslip. Fluorescence imaging was carried out on an iMIC widefield microscope (FEI) with a 40x oil objective and an Hamamatsu Orca camera. Z stacks were acquired with a slice separation of 1 μm and deconvolved using SVI Huygens before maximal intensity projection along the z-axis.
Cloning of gpa-4 Cre driver
The plasmid used in this study was generated using Gibson assembly. The gpa-4 promoter was amplified from plasmid pLSD146, in which it drives expression of GFP (Jansen et al. 1999) using primers 1581/1582. The degron sequence was amplified from plasmid #503 using primers 1583/1584. The rest of the plasmid which contains the Cre::tbb-2 3’UTR sequence and the backbone pCFJ355 (Frøkjær-Jensen et al. 2012), including homology regions to the ttTi14024 Mos insertion were amplified separately in two pieces using primers 1577/1578 and 1579/1580 and the plasmid pSR33 as a template (Ruijtenberg and van den Heuvel 2015). All PCR reactions were then digested with DpnI and fragments ligated together using Gibson assembly. The Cre ORF ATG in the new plasmid is located at the same place relative to the gpa-4 promoter than in the original pLSD146 plasmid.
Reagents
Primer collection.
| Primer name | Sequence | Template |
| PM1581 | GAGGGTACCAGAGCTCACCTAGGTATCTAGCAAGCTTGCGACTTTCGATACGTAGGTCTC | pLSD146 |
| PM1582 | CTTATTCATTTTGTGAACACTTTTCAACAAAGCTTGATGCCTAAAGATCCAGCCAAACCTCCGGCC | pLSD146 |
| PM1583 | CATTTTGTGAACACTTTTCAACAAAGCTTGATGCCTAAAGATCCAGCCAAACCTCCGGCC | #503 |
| PM1584 | GTAGTCTCCATCGTGATCCTTGTAATCCATCTTCACGAACGCCGCCGCCTCCGGGCC | #503 |
| PM1577 | ATGGATTACAAGGATCACGATGGAGACTACAAGG | #328 |
| PM1578 | CCCTGGCGTTACCCAACTTAATCGCCTTGC | pSR33 |
| PM1579 | GCAAGGCGATTAAGTTGGGTAACGCCAGGG | pSR33 |
| PM1580 | GCTAGATACCTAGGTGAGCTCTGGTACCCTCTAG | pSR33 |
| PM1323 | CCTCGTTTCGAAGTTGGTTTG | gDNA |
| PM1324 | CTCCGGTGAAGAGCTCCTCTCC | gDNA |
Plasmids
| Plasmid name | Source | Plasmid information | Backbone |
| #522 | This study | gpa-4p::Degron::Cre::tbb-2 3’UTR | pCFJ355 |
| pLSD146 | Jansen et al.1999 | gpa-4p::gfp | – |
| #503 | Gomez, Osuna-Luque et al., 2020 | hsp16.2p::lox::mCherry::lox::degron::Dam::rpb-6::unc54 3’UTR | pCFJ151 |
| pSR33 | Ruijtenberg and van der Heuvel, 2015 | hsp16.2p::Cre::tbb-2 3’UTR | pCFJ355 |
C. elegans strains
Plasmid #522 was integrated as a single copy using MosSCI on the chromosome X ttTi14024 Mos insertion site (Frøkjaer-Jensen et al. 2008).
| Strain name | Source | Genotype |
| N2 | CGC | Wild type Bristol N2 |
| PMW848 | This study | ubsSi45[gpa-4p::degron::Cre::tbb-2 3’UTR; unc-119(+) @ttTi14024] X |
| SV1361 | Ruijtenberg and van der Heuvel, 2015, CGC | heIs105 [rps-27::loxP::nls::mCherry::let858 3’UTR::loxP::nls::GFP::let-858 3’UTR; unc-119(+) @cxTi10816] IV |
| SV1439 | Ruijtenberg and van der Heuvel, 2015, CGC | heSi142[elt-2p::egl-13nls::Cre::tbb-2 3’UTR; unc-119(+) @ttTi14024] X |
| PMW417 | This study | heSi104[rps-27p::loxP::nls::mCh::let-858 3’UTR::loxP::nls::gfp::let-858 3’UTR;unc-119(+) @cxTi10816] IV; unc-119(?) III; heSi142[elt-2p::egl-13nls::Cre::tbb-2 3’UTR @ttTi14042] X |
| PMW867 | This study | heIs105 [rps-27::loxP::nls::mCherry::let858 3’UTR::loxP::nls::GFP::let-858 3’UTR; unc-119(+) @cxTi10816] IV; ubsSi45[gpa-4p::degron::Cre::tbb-2 3’UTR; unc-119(+) @ttTi14024] X |
Table 1. gpa-4 promoter-driven gene expression studies
| First author | PMID | gpa-4 promoter driving expression of |
| Gallagher et al. | 23739968 | daf-11, egl-4, GCaMP2.2b |
| Guo et al. | 30428355 | R-GECO1 (red protein), TRPV1, TeTx, ser-3 |
| Cheong-Cheong et al. | 25898004 | npr-17::SL2::RFP |
| Broekhuis et al. | 23444385 | sql-1::GFP, aman-2::YFP |
| Murakami et al. | 11677050 | daf-11 cDNA |
| Chen et al. | 27585848 | NLS::mCherry, mouse OxR2 |
| Pandey et al. | 33712439 | exp-1::SL2::wrmScarlet, str-2::SL2::wrmScarlet |
| Kim et al. | 16143323 | unc-3, GFP |
| Dixit et al. | 32432390 | str-2 |
| You et al. | 18316030 | daf-7 |
| Bishop et al. | 17538612 | skn-1b::gfp |
Acknowledgments
We thank Gert Jansen for the gpa-4p::gfp plasmid.
References
Funding
Funding from the Swiss National Science Foundation PP00P3_190072 to CYE and IZCOZ0_189884/31003A_176226 to PM.
Reviewed By
AnonymousHistory
Received: June 3, 2021Revision received: August 3, 2021
Accepted: August 5, 2021
Published: August 19, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Osuna-Luque, J; Ewald, CY; Meister, P (2021). Widespread gpa-4 promoter-driven expression during Caenorhabditis elegans development. microPublication Biology. 10.17912/micropub.biology.000443.Download: RIS BibTeX