Abstract
Mitochondria are ATP-producing organelles that also signal throughout the cell. Mitochondrial protein homeostasis is regulated through membrane potential-dependent protein import and quality control signaling. The mitochondrial unfolded protein response (UPRmt) is a specific program that responds to imbalances in nuclear and mitochondrial gene expression. Mounting evidence suggests that the electrochemical gradient that powers mitochondrial function, the mitochondrial membrane potential (Δψm), is a core regulator of the UPRmt. Here we tested this notion directly by pharmacologically dissipating Δψm and monitoring UPRmt activation. We found that chemical dissipation of Δψm using FCCP indeed activated UPRmt dose-dependently in C. elegans assayed by the HSP-60::GFP reporter strain.
Description
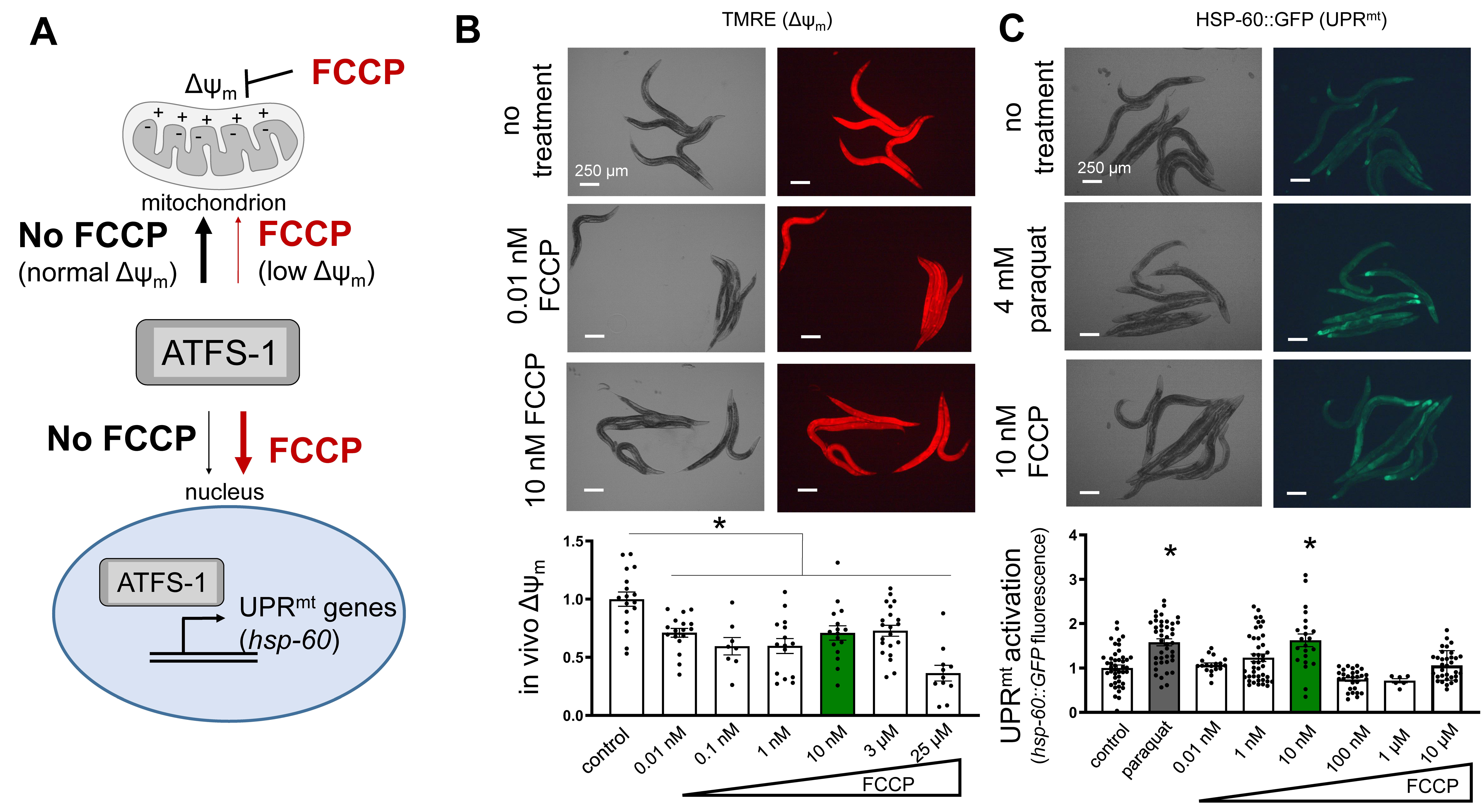
Mitochondria are organelles that make ATP by generating a proton gradient across the inner membrane through the action of the electron transport chain (ETC). The gradient is composed of a membrane potential (Δψm) and a concentration gradient (ΔpH). In addition to energy production, mitochondria are recognized as signaling organelles (Chandel 2015), and Δψm powers diverse signaling outputs that mitochondria coordinate. One such signaling response is the mitochondrial unfolded protein response (UPRmt). Some ETC complex components are encoded by mitochondrial or nuclear genes and require coordination between the genomes for proper stoichiometry (Shpilka and Haynes 2018). The UPRmt is a protein homeostasis response that is activated when nuclear and mitochondrial gene expression are not correctly coordinated (Melber and Haynes 2018). When the expression of mitochondrial and nuclear encoded proteins is mismatched, the UPRmt is activated. In C. elegans, the UPRmt is controlled by the transcription factor ATFS-1, which is normally trafficked to mitochondria and degraded. When nuclear:mitochondrial gene expression is perturbed, ATFS-1 is blocked from entering mitochondria and is instead trafficked to the nucleus where a host of chaperones and stress-response genes are activated and transcribed.
Recently, it was suggested that ATFS-1 entry to mitochondria for degradation depended on sufficient Δψm (Rolland et al. 2019). In vivo evidence supported this assertion, and implied that Δψm is a main regulator of mitochondrial protein homeostasis through its role in driving ATFS-1 import to mitochondria (Poveda-Huertes et al. 2021; Shpilka et al. 2021). Based on this premise, we sought to directly test if targeting Δψm could alter UPRmt signaling. We predicted that decreased Δψm would result in activation of UPRmt due to decreased driving force for ATFS-1 to enter mitochondria and be degraded. This would lead to ATFS-1 localization to the nucleus and activation of the UPRmt target gene hsp-60 (Figure 1A). We used the protonophore FCCP at various doses to decrease Δψm in live C. elegans. Δψm was monitored in live, adult worms using the potentiometric fluorescent indicator, TMRE. At all doses tested Δψm was decreased compared to untreated control (Figure 1B). Using a similar dose range, we found that one intermediate dose was sufficient to activate UPRmt signaling, assayed through the characterized transcriptional hsp-60 reporter strain (Melber and Haynes 2018; Rolland et al. 2019) (Figure 1C). Paraquat, a known UPRmt positive control (Kim and Sieburth 2018), was used to confirm our results with FCCP. This study was repeated on at least 3 independent days.
The UPRmt is linked to many stress-resistance signaling responses, including hypoxia resistance (Peña et al. 2016). We previously showed that FCCP protected against hypoxia (Berry et al. 2020b), and this work suggests that UPRmt may have been involved. Our other work, however, implicated energy-sensing signaling downstream of decreased Δψm and hypoxia resistance (Berry et al. 2020a). The requirement of UPRmt for hypoxia resistance downstream of decreased Δψm remains untested. Further, the UPRmt has been implicated in many disease models that are not fully characterized (Jovaisaite et al. 2014). It is likely that many changes occur with decreased Δψm, and it will be necessary to characterize changes in Δψm and other mitochondrial functions in a wide range of contexts to uncover molecular mechanisms of mitochondrial signaling. For example, it is unclear why only a certain dose (10 nM) of FCCP activated the UPRmt here when all doses resulted in mitochondrial depolarization. This result suggests that other cellular mechanisms likely control UPRmt activation in addition to Δψm. In addition, the level of mitochondrial depolarization may differentially result in UPRmt or mitochondrial autophagy, a process that is also responsive to decreased Δψm. The mechanism that controls which cellular response is elicited likely depends on the degree of mitochondrial stress. This work is complementary to the established role of mitochondrial ETC stress and UPRmt. Inhibition of the ETC results in activation of UPRmt, but only if initiated before the L3/L4 stage in C. elegans (Durieux et al. 2011). Our results suggest that the mechanism of ETC mediated UPRmt activation may be somewhat distinct from the Δψm control over UPRmt, because decreased Δψm was able to induce UPRmt after the L3/L4 stage.
Methods
Request a detailed protocolFluorescence imaging. C. elegans were fed OP50 bacteria on nematode growth media (NGM) plates. L4 animals were synchronized by egg lay and were stained by exposure to 100 nM TMRE for 24 hours that was applied to plates (Cho et al. 2017; Berry et al. 2020B). Fluorescence was photographed in day 1 adult animals on 2% agarose pads with tetramisole (0.1% w/v) for anesthetic. Red TMRE fluorescence was captured using TexasRed filter sets, and green GFP was captured using GFP filter sets on an epifluorescence microscope (Olympus MVX) equipped with a Lumenera camera and acquisition software (Infinity Analyze; Lumenera). Fluorescence of individual animals was quantified using ImageJ. ROIs were whole animals, and background was manually subtracted from final measurements in ImageJ. Mean intensity was measured, and after background subtraction, was normalized to control intensity. Paraquat was dissolved in water and applied to plates at a final concentration of 4 mM. FCCP was dissolved in ethanol (< 0.1% final concentration, consistent across all concentrations) and applied to plates at various concentrations (see figure). Animals were exposed to paraquat or FCCP for 24 hours.
Reagents
| Strain | Genotype | Source |
| N2 | wildtype | CGC |
| SJ4058 | zcIs9 [hsp-60::GFP + lin-15(+)] V | CGC |
Acknowledgments
B.J.B. current affiliation: Department of Laboratory Medicine and Pathology, University of Washington Medical Center, 1959 NE Pacific St, Seattle, WA, 98195, USA.
References
Funding
A.P.W. is supported the National Institutes of Health (R01 NS092558, R01 NS115906). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
AnonymousHistory
Received: July 21, 2021Revision received: August 9, 2021
Accepted: August 21, 2021
Published: September 10, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Berry, BJ; Nieves, TO; Wojtovich, AP (2021). Decreased Mitochondrial Membrane Potential Activates the Mitochondrial Unfolded Protein Response. microPublication Biology. 10.17912/micropub.biology.000445.Download: RIS BibTeX