Abstract
Histone modifications influence gene expression and chromosome dynamics by altering chromatin structure and recruitment of nonhistone proteins. Dimethylation of histone H3 on lysine 9 (H3K9me2) is a conserved modification often found within heterochromatin. During first meiotic prophase when homologous chromosomes undergo pairing and synapsis, immunolabeling of C. elegans male germ cells detects a relatively high H3K9me2 level on the single X chromosome and a relatively low H3K9me2 level on synapsed autosomes. This H3K9me2 distribution is influenced by several components of the small RNA machinery, including: EGO-1 RNA-directed RNA polymerase (RdRP); DRH-3 helicase; EKL-1, a Tudor domain protein; CSR-1 Argonaute; and RRF-3 RdRP. EGO-1, DRH-3, and EKL-1 function together to generate/stabilize 22G RNAs in the germ line. A subset of these 22G RNAs function together with CSR-1 to ensure correct gene expression. RRF-3 RdRP functions in biogenesis of 26G RNAs that feed into two germline regulatory mechanisms mediated by ERGO-1 Argonaute and the redundant ALG-3 and ALG-4 Argonaute proteins. Here, we report that meiotic H3K9me2 distribution is influenced by ALG-3 and ALG-4, as well as by two other factors required for 26G RNA synthesis, ERI-1 and ERI-5. Moreover, meiotic H3K9me2 distribution is influenced by activity of the poly(U) polymerases, PUP-1 (aka CDE-1, CID-1) and PUP-2.
Description
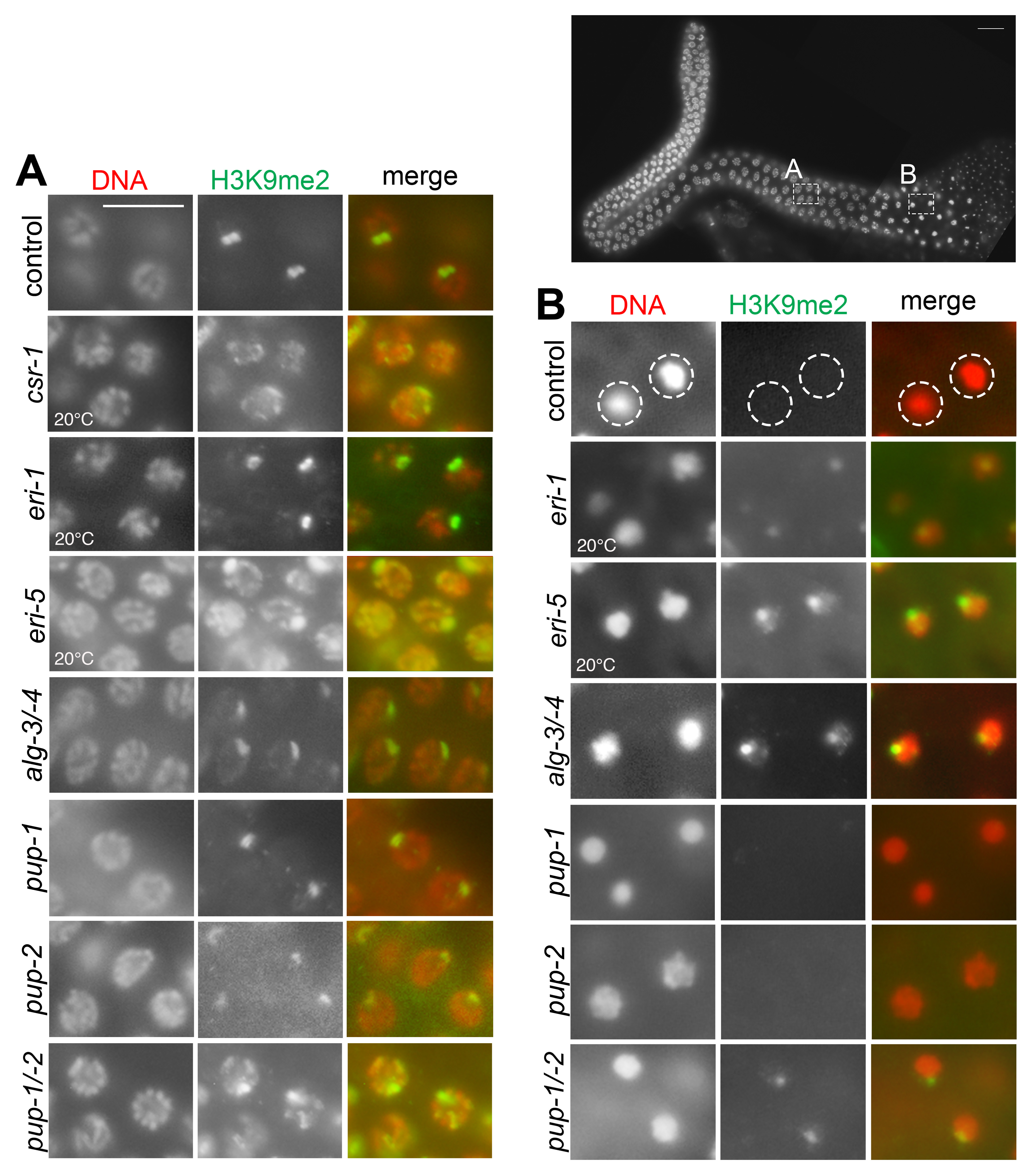
Histone modifications are precisely controlled within the developing germ line. During first meiotic prophase in C. elegans, H3K9me2 immunolabeling detects a relatively bright focus of signal on unsynapsed chromosomes and a relatively weak, diffuse signal on synapsed chromosomes (Bean et al. 2004, Kelly et al. 2002, Reuben and Lin 2002). The naturally occurring unsynapsed chromosome is the single male X. Examples of other chromatin that is enriched for H3K9me2 at pachytene stage include single-copy free chromosomal duplications, chromosomes that fail to pair due to certain mutations, and multicopy extrachromosomal arrays. In all cases, the enriched H3K9me2 signal decreases sharply as nuclei move into diplotene stage, and H3K9me2 is not detected in diakinesis nuclei. As the naturally occurring example of H3K9me2 enrichment, the male X is commonly used to evaluate this meiotic targeting phenomenon (Fig. 1). We previously showed that meiotic H3K9me2 distribution is altered in males lacking CSR-1 Argonaute or factors required for accumulation of CSR-1-associated 22G RNAs, including EGO-1 RdRP, DRH-3 helicase, and the Tudor domain protein, EKL-1 (Maine et al. 2005, She et al. 2009). In mutants lacking any of these factors, pachytene H3K9me2 signal intensity is reduced on unsynapsed chromosomes and, in all cases except ego-1(null) mutants, ectopic H3K9me2 foci are detected on synapsed chromosomes. Interestingly, as in wildtype, H3K9me2 signal is not detected in diakinesis nuclei in these mutants. CSR-1 pathway activity is essential for germ line development where it is thought to license the correct pattern of gene expression (Yigit et al. 2006, Ashe et al. 2012, Shirayama et al. 2012). EGO-1, DRH-3, and EKL-1 are critical for biogenesis and stability of 22G RNAs that associate with CSR-1 (reviewed in Billi et al. 2014).
RRF-3 RdRP is essential for biogenesis of 26G RNAs that participate in the ALG-3 and ALG-4 Argonaute pathway during spermatogenesis and the ERGO-1 Argonaute pathway during oogenesis (Billi et al. 2014). We previously observed an essentially wildtype H3K9me2 immunolabeling pattern in rrf-3(pk1426) males during first meiotic prophase that then persists (presumably on the X chromosome) beyond diplotene and is visible on karyosomes in the late spermatogenesis condensation zone as described by Shakes et al. (2009) (Maine et al. 2005). We observed wildtype H3K9me2 distribution in ergo-1(tm1860) single mutant, alg-3(tm1155) single mutant, and alg-4(ok1041) single mutant males (She et al. 2009), but we did not test alg-4(ok1041);alg-3(tm1155) double mutants at that time. We subsequently hypothesized that the rrf-3(pk1426) H3K9me2 phenotype may reflect impaired ALG-3 and ALG-4 pathway activity, since these Argonaute proteins together are critical during spermatogenesis (Han et al. 2009, Conine et al. 2010, 2013) at the time when the H3K9me2 signal typically decreases.
Here, we follow up on our earlier results to report meiotic H3K9me2 distribution in eri-1(mg366) single mutant, eri-5(tm2528) single mutant, and alg-4(ok1041);alg-3(tm1155) double mutant males. ERI-1, an exoribonuclease, and ERI-5, a Tudor domain protein, are both required for 26G RNA synthesis. We also evaluated H3K9me2 distribution in pup-1(tm1021) single mutant, pup-2(tm4344) single mutant, and pup-1/-2(om129) double mutant males. PUP-1 (aka CDE-1, CID-1) and PUP-2 are members of the poly(U) polymerase family responsible for adding non-templated uridine(s) to the 3’ end of RNA. PUP-1 is implicated in adding 3’ uridine to 22G RNAs that associate with CSR-1 and WAGO-4 Argonautes (van Wolfswinkel et al. 2009, Xu et al. 2018) and to microRNAs (Vieux et al. bioRxiv preprint). PUP-2 promotes germline development redundantly with PUP-1 under conditions of heat stress (Li and Maine 2018) and has been implicated in regulating stability of let-7 miRNA (Lehrbach et al. 2009). U-tailing of small RNAs may influence their stability and/or Argonaute associations, as has been suggested (Billi et al. 2014, Xu et al. 2018).
We immunolabeled H3K9me2 in adult him-8 control males raised at 20°C and 25°C and in various mutant backgrounds. We tested alg-4(ok1041);alg-3(tm1155) double mutant, eri-1(mg366) single mutant, eri-5(tm2528) single mutant, pup-1(tm1021) single mutant, pup-2(tm4344) single mutant, and pup-1/-2(om129) double mutant males. The alg-4(ok1041);alg-3(tm1155) double mutant, pup-1(tm1021) single mutant, pup-2(tm4344) single mutant, and pup-1/-2(om129) double mutant phenotypes are all temperature sensitive, and these mutants were therefore tested at restrictive temperature (25°C). The pup single and double mutants were F1 (M+Z-) offspring (where M indicates maternal genotype and Z indicates embryonic genotype) of heterozygous hermaphrodites (see Methods). We note that pup-1/-2(om129) M+Z- males all produce sperm, although many of those sperm have abnormal morphology and are fertilization defective (Li and Maine 2018). H3K9me2 immunolabeling in pachytene nuclei appears wildtype in eri-1(mg366) single mutant, alg-4(ok1041);alg-3(tm1155) double mutant, pup-1(tm1021) single mutant, and pup-2(tm4344) single mutant males; in each case, a strong signal is visible in one region of the genome, and weak, diffuse signal is visible elsewhere across the genome (Fig. 1A). In contrast, multiple discrete H3K9me2 foci are visible in pachytene nuclei of pup-1/-2(om129) M+Z- double mutant and eri-5(tm2528) single mutant males (Fig. 1A). In spermatogenic nuclei, H3K9me2 is not detected in pup-1 or pup-2 single mutants, as it is not in the control (Fig. 1B). In contrast, H3K9me2 is detected in karyosomes in alg-4(ok1041);alg-3(tm1155) double mutant, eri-1(mg366) single mutant, eri-5(tm2528) single mutant, and pup-1/-2(om129) M+Z- double mutant males (Fig. 1B). We note that H3K9me2 appears wildtype pup-1/-2(om129) F1 (M+Z-) double mutants at 20°C, consistent with the temperature-sensitivity of other aspects of the pup phenotype (Spracklin et al. 2017, Li and Maine 2018).
Our working model is that the delayed H3K9me2 turnover in rrf-3(pk1426) single mutant, alg-4(ok1041);alg-3(tm1155) double mutant, eri-1(mg366) single mutant, and eri-5(tm2528) single mutant males reflects a role for the ALG-3 and ALG-4 pathway in chromatin regulation during spermatogenesis. The broader H3K9me2 phenotype in pup-1/-2(om129) M+Z- 25°C mutants may reflect impaired activity of both the CSR-1 pathway and the ALG-3 and ALG-4 pathway, perhaps due to misrouting of siRNAs that would typically associate with these Argonaute proteins. CSR-1 promotes spermatogenesis, and it is implicated as functioning downstream of ALG-3 and ALG-4 to regulate gene expression during spermatogenesis (Conine et al. 2010, 2013; Charlesworth et al. 2021) although CSR-1 does not appear to act directly downstream of ALG-3 and ALG-4 in all cases (Nguyen and Phillips 2021). Misrouting of siRNAs may occur, at least in part, due to absence of 3’ U-tailing as has been proposed by van Wolfswinkel et al.. (2009), Xu et al. (2018), and others.
Methods
Request a detailed protocolImmunocytochemistry and DAPI staining: Gonads were dissected from L4 + 24 hr adults and immunolabeled with anti-H3K9me2 antibody as described (Maine et al. 2005; She et al. 2009). Tissue was fixed in 3% PFA/PBS solution for 5 min. Mouse anti-H3K9me2 (Abcam 1220) primary antibody and Alexa Fluor 488-conjugated goat anti-mouse secondary antibody were each used at a 1:200 dilution. Tissue was stained with DAPI in the penultimate wash. Slides were observed with a Zeiss Axioscope or Leica DM5500 microscope. Images of H3K9me2 immunolabeling in different genetic backgrounds were taken at the same exposure.
Reagents
Strains used in this study:
| Strain | Genotype & Linkage Group | Notes |
| CB1489 | him-8(e1489) LG IV | |
| EL502 | eri-1(mg366) him-8(e1489) LG IV | |
| EL567 | alg-4(ok1041);alg-3(tm1155) LG III;IV | Temperature-sensitive phenotype. Mating population was maintained at 20°C. |
| EL609 | csr-1(om135) him-8(e1489)/mIs11 him-8(om133) LGIV | Strain construction is described below. |
| EL624 | pup-1/-2(om129)/qC1gfp;him-8(e1489) LG III;IV | om129 deletes the adjacent pup-1 and pup-2 genes. Temperature-sensitive phenotype. |
| EL625 | pup-2(tm4344);him-8(e1489) LG III;IV | Temperature-sensitive phenotype. |
| EL690 | pup-1(tm1021)/qC1gfp;him-8(e1489) LG III; IV | Temperature-sensitive phenotype. |
| FX2528 | eri-5(tm2528) LG IV | Maintained as a mating population. |
| PD4792 | mIs11[myo-2p::GFP + pes10p::GFP + gut-promoter::GFP] LG IV |
We used CRISPR-Cas9 editing to generate csr-1(om135) in the CB1489 him-8(e1489) strain background and to generate him-8(om133) in the PD4792 mIs11 balancer strain background. For each mutation, we injected sets of sgRNAs designed to delete a large portion of the gene. csr-1(om135) contains a deletion with breakpoints in exon 1 and 11. The deletion removes 3530 nucleotides corresponding to (1) most of the coding region for the longer CSR-1A isoform and (2) the start codon plus most of the coding region for the shorter CSR-1B isoform. The sterile phenotype is 100% penetrant. him-8(om133) contains a deletion with breakpoints in exon 4 and 6, removing the 3’ portion of the coding region. The Him phenotype is 100% penetrant.
Acknowledgments
We thank Bing Yang for DNA injections to produce CRISPR-generated alleles and Xingyu She for preliminary H3K9me2 immunolabeling of eri-1 mutant and 25°C wildtype strains (PhD dissertation 2010). We thank Sarah Hall, Leanne Kelley, and an anonymous reviewer for comments on the manuscript, and members of the Maine and Hall Labs for discussions. Some strains used in this study contain mutations obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, and/or the National BioResource Project under the auspices of Shohei Mitani.
References
Funding
This work was supported by National Institutes of Health grant R01GM089818.
Reviewed By
AnonymousHistory
Received: August 23, 2021Revision received: September 2, 2021
Accepted: September 6, 2021
Published: September 14, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Li, Y; Snyder, M; Maine, EM (2021). Meiotic H3K9me2 distribution is influenced by the ALG-3 and ALG-4 pathway and by poly(U) polymerase activity. microPublication Biology. 10.17912/micropub.biology.000455.Download: RIS BibTeX