Northeastern University, Department of Bioengineering, Boston, MA, USA
Baylor University, Department of Biology, Waco, TX, USA
Abstract
In C. elegans, oocytes are ovulated into the spermatheca, where they are fertilized before being pushed into the uterus. Contraction in the C. elegans spermatheca is driven by circumferential acto-myosin fibers. The C. elegans zyxin homolog, zyx-1, is expressed in the body wall muscle, pharynx and spermatheca. To our surprise, a CRISPR-generated zyx-1 deletion allele results in no overt developmental phenotypes, and the spermathecal actin cytoskeleton appears wild type, however, oocyte transit through the spermatheca is slower than in wild type animals. This suggests ZYX-1/Zyxin may regulate spermathecal contraction magnitude or timing of spermathecal bag contraction and/or spermathecal-uterine valve dilation.
Description
Zyxin is a LIN-11, Isl1, and MEC-3 (LIM)-domain protein that, in mammalian cells, binds alpha actinin and other cytoskeletal-associated proteins. Zyxin localizes to the integrin-based adhesive structures known as focal adhesions and to stress fibers, which are contractile acto-myosin bundles that regulate cell shape and contractility (Wang et al. 2019). The C. elegans protein ZYX-1, a protein with similarity to zyxin, migfilin, TRIP6, and LPP (Lecroisey et al. 2013), was identified in a screen for binding partners of germline RNA helicase (GLH) proteins (P. Smith et al. 2002). Similar to mammalian zyxin, ZYX-1 contains three LIM domains, a potential nuclear export signal (NES) and three proline rich regions (PRR). ZYX-1 is localized to the dense body/Z-disk in body wall muscle and interacts with DEB-1/vinculin (Lecroisey et al. 2013), ATN-1/α-actinin (Lecroisey et al. 2013) , and DYC-1, a dense-body specific protein that potentially interacts with DYS-1/dystrophin (Lecroisey et al. 2008). ZYX-1 might play a role in signaling between the cytoplasm and the nucleus, as a portion of the ZYX-1 protein is translocated from the cytoplasm to the nucleus (Lecroisey et al. 2013). Mounting evidence in mammalian cells suggests that zyxin is mechanosensitive and re-localizes to stress fibers that are under tension (Wang et al. 2019; M. Smith et al. 2010; Schiller et al. 2011), and participates in the repair of damaged actin fibers (M. Smith et al. 2010). It is not known whether ZYX-1 fulfills a similar function in C. elegans.
The C. elegans spermatheca is a contractile tube of smooth muscle-like cells found in the hermaphroditic reproductive system. Oocytes are ovulated into the spermatheca, where they are fertilized. The spermatheca then contracts to push the fertilized embryos out through the spermathecal-uterine (sp-ut) valve and into the uterus (Laband et al. 2018; Yamamoto, Kosinski, and Greenstein 2006). Oocyte entry stretches the cells of the spermatheca and leads to the formation of basal acto-myosin bundles. These stress fiber-like bundles drive spermathecal contractility (Wirshing and Cram 2017). However, while the spermatheca expresses integin and vinculin, dense bodies are not apparent in this tissue (Ono, Yu, and Ono 2007) and how basal acto-myosin bundles are anchored in this tissue is not well understood. Because of the strain placed on these fibers by embryo transit, we asked if the C. elegans homolog of zyxin, ZYX-1, might play a role in assembly or maintenance of the actin cytoskeleton in the spermatheca.
In this study, we investigated the role of C. elegans ZYX-1/Zyxin in spermathecal contractility and regulation of the actin cytoskeleton using a novel CRISPR-generated deletion allele. The animals are superficially wildtype and do not exhibit overt cytoskeletal defects. However, when zyx-1 is disrupted, oocyte transit through the spermatheca is slower than in wild type animals. This suggests ZYX-1/Zyxin may regulate the magnitude or timing of spermathecal contractility.
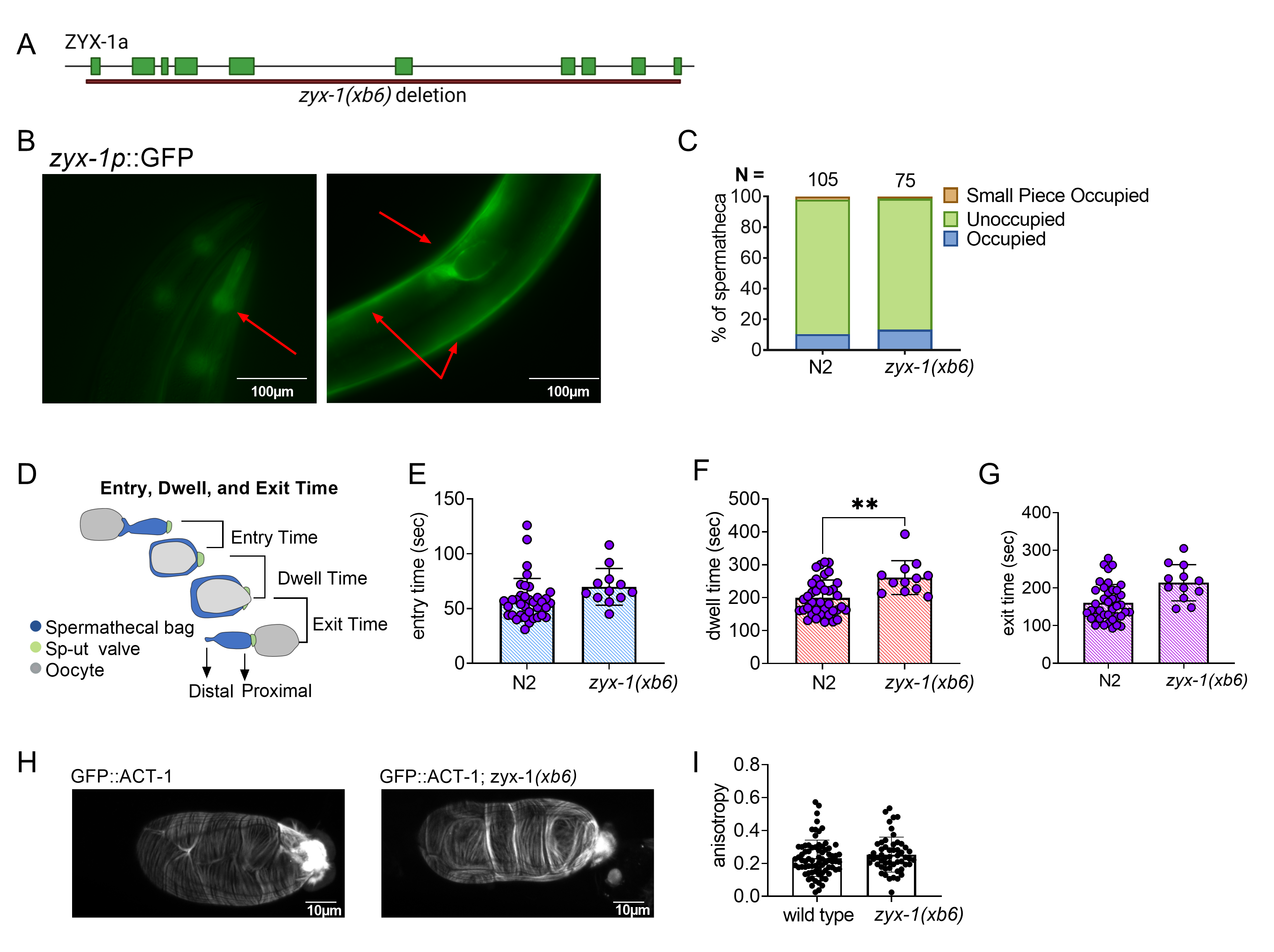
To characterize the function of ZYX-1/Zyxin in C. elegans, we deleted zyx-1 using CRISPR genome editing. The resulting deletion allele, zyx-1(xb6), is a 13,503 bp deletion with a 12 bp insertion, which removes the entire coding sequence of F24G4.3a (Fig. 1A). The zyx-1(xb6) animals were backcrossed four times to N2 to remove off-target mutations, and the deletion was verified by PCR and sequencing. The deletion animals did not exhibit any overt developmental, movement, or growth phenotypes.
We next investigated the expression pattern of ZYX-1/Zyxin using BC14184, a strain expressing a transcriptional fusion of ZYX-1::GFP. ZYX-1::GFP is expressed in the pharynx, body wall muscles, and spermatheca of C. elegans (Fig. 1B). Because ZYX-1 is strongly expressed in the spermatheca, we asked if ZYX-1 regulates embryo transits through the spermatheca. Age matched, young adult zyx-1(xb6) and N2 animals were scored for spermathecal occupancy using differential interference contrast (DIC) microscopy. No statistically significant increase in spermathecal occupancy was observed (Fig. 1C). While a small piece of an oocyte was observed in the zyx-1(xb6) worms, this was also observed in our N2 control.
To detect subtle defects in spermathecal transits, we used video microscopy to record ovulations and transits of zyx-1(xb6) and N2 animals and calculated the entry time, dwell time and exit time. Entry time is the time from the opening of the distal spermatheca to the complete entry of the oocyte and closure of the distal spermathecal neck. Dwell time is the time when the embryo is completely enclosed by the spermatheca. Exit time is the time from the opening of the sp-ut valve to the complete expulsion of the embryo into the uterus (Fig. 1D). We observed little difference between the two genotypes in terms of entry time and exit time, but the dwell time was significantly longer in the zyx-1 deletion than in the wild type (Fig. 1E-G). This suggests that ZYX-1/Zyxin promotes embryo transit through the spermatheca.
In cell culture systems, zyxin is required to maintain stress fibers (M. Smith et al. 2010), especially under tension. We speculated that defects in the actin cytoskeleton might explain the observed zxy-1(xb6) transit phenotype. To determine if ZYX-1/Zyxin regulates spermathecal actin, we crossed zxy-1(xb6) into a strain expressing GFP::ACT-1 and observed spermathecal transits using confocal microscopy. However, no difference was observed in the actin fiber alignment in wild type and zyx-1(xb6) spermathecae (Fig. 1H, I).
Here, we demonstrate that ZYX-1/Zyxin is not required per se for spermathecal transits or for actin alignment in the spermatheca. ZYX-1/Zyxin does seem to be involved in regulation of the magnitude or timing of spermathecal contractility in C. elegans, however, the mechanism of this regulation remains elusive.
Why might zyx-1(xb6) have such mild phenotypes? ZYX-1/Zyxin may only become necessary when the animals are stressed by loss of another regulator. For example, depletion of DYS-1/Dystrophin and ZYX-1/Zyxin in body wall muscle (Lecroisey et al. 2008) leads to defects in the actin cytoskeleton and paralysis. Although ZYX-1 is the sole zyxin/migfilin/TRIP6/LPP (Lecroisey et al. 2013) homolog in C. elegans, other actin associated proteins may serve functionally redundant roles in regulation of the actin cytoskeleton and spermathecal contractility. Future proteomic and/or genetic studies may lead to a better understanding of the role ZYX-1/Zyxin in C. elegans.
Methods
Request a detailed protocolC. elegans strains and culture
C. elegans were maintained at 23°C, on NGM agar plates ((0.107 M NaCl, 0.25% wt/vol Peptone, 1.7% wt/vol BD Bacto‐Agar, 2.5 mM KPO4, 0.5% Nystatin, 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5% wt/vol cholesterol) seeded with E. coli OP50. C. elegans BC14184 (ZYX-1::GFP) and N2 strains were obtained from the C. elegans Genetics Center.
To delete the RGD motif in zyx-1 locus in chromosome II, we identified two effective CRISPR sites to delete about 10 kb from exon 1 to 10 from the CRISPR guide RNA Selection Tool, http://genome.sfu.ca/crispr/search.html. According to the intended mutation, 84-mer repair DNA templates containing 25-base homology arms (zyx-1 null, see the reagent section) were designed and custom-made by IDT Inc., Coralville, IA. Then, the mixture of template DNA (ZYX-1 template name), two crRNA (zyx1start and zyx1stop, see the reagent section), tracrRNA (catalog no. 1072532), and Alt-R Cas9 (catalog no. 1081058) proteins are annealed at 37oC and micro-injected into the syncytial gonad arms of N2 animals (P0) with dpy-10 crRNA as a co-CRISPR marker (Paix et al. 2015; Dickinson and Goldstein 2016). The F1 offspring of P0 worms are selected by Dpy and subjected to PCR genotyping to identify worms carrying the desired deletions. Once the F1 mutants are isolated, F2 progeny are screened to identify the homozygous alleles as described below.
PCR was used to verify the genomic modification. Briefly, single animals were lysate in 6 μl worm lysis buffer (10 mM Tris HCl pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.45% Tween 20, 0.45% Triton X100) and PK (Proteinase K) and frozen at -80℃. After protease digestion at at 60℃ for 1 hr and denaturation at 95℃ for 15 min, the lysate was used for PCR with GoTaq green master mix using the primer pairs, ZYX-1 null Forward (TGTCTTTCAGCTTGGGTCGT) and ZYX-1 null Reverse (GGTTGGCATCCGTACTCGAA) to amplify the zyx-1 locus. The edit was verified by sequencing. Isolated PCR products were sequenced to confirm the mutations. The established homozygous zyx-1 deletion lines were backcrossed to N2 several times to reduce off-target effects. The resulting strain, UN2013 (zyx-1(xb6)), was crossed with UN1502 (GFP::ACT-1) to produce UN2016 (Y66H1Bp::GFP::ACT-1; zyx-1(xb6)).
Preparation of populations of young adult hermaphrodites
To obtain age-matched young adult hermaphrodites, dauer nematodes were placed on fresh, seeded NGM plates and raised for 48 hours. Eggs were harvested from gravid adults using alkaline lysis as described (Hope 1999), and were left to starve overnight to arrest at L1. L1 animals were raised in a 23°C incubator and observed after 50-54 hours.
DIC, fluorescence and confocal microscopy
Animals were mounted on 5% agarose in 0.1 M sodium azide and observed immediately with DIC microscopy to score gonad phenotypes. Imaging was performed on a Nikon Eclipse 80i microscope with a 60× oil-immersion lens using SPOT Advanced software (Version 5.3.5) and a charge-coupled device camera. Frames were captured at a rate of 1 Hz. Image stacks were reassembled and analyzed using ImageJ software (Schindelin et al. 2012). Fluorescence microscopy was performed on a Nikon Eclipse 80i microscope equipped for epifluorescence. Images were captured with a SPOT RT3 CCD camera using SPOT Advanced software (Diagnostic Instruments; Sterling Heights, MI, USA). Confocal microscopy of animals expressing GFP::ACT-1 was performed on a Zeiss LSM 710 confocal using Zen software and a Plan-Apochromat 63x/1.40 oil objective lens using a 488-nm laser (Kelley et al. 2020).
Quantification with ImageJ
We used ImageJ software (version 2.1.0/1.53c) to analyze the ovulation movies and to measure the length of the worms. Open ImageJ, right-click on the ‘straight line’ tool, select ‘freehand line’, draw a line along the shape of the worms, and press command+M to get the pixel value. The length of worms is calculated from the pixel value through a conversion based on the calibrated scale bar.
Analysis of actin alignment with Fibriltool, an ImageJ plugin
Fibriltool was used essentially as described in Boudaoud, A et al., (Boudaoud et al. 2014) Briefly, the steps are to open the file, split the channels, apply z project, and process and save the image in PNG format (a key step for success), before following the steps in the article.
Statistics
All of the statistical analysis was performed using GraphPad Prism (version 9.2.0). For comparing transit timepoints and anisotropy, the Student’s t-test was used. The Fisher’s Exact test was used to compare proportions of occupied spermatheca.
Reagents
| Oligo Name | Sequence (5’-3’) |
| ZYX1NULLF
(PCR genotyping) |
TGTCTTTCAGCTTGGGTCGT |
| ZYX1NULLR
(PCR genotyping) |
GGTTGGCATCCGTACTCGAA |
| ZYX1WTR
(PCR genotyping) |
ATCCCTGAGCTTTTGGGGGT |
| zyx-1 null REPAIR TEMPLATE (84 bases) | ggggggaatggaaattgttgactgatggctcgcTTGCTAGCGCTAGCtagcggatgccgagtctggaatagtgctgaaggag |
| zyx1start (crRNA) | GGGGTCCCATagatcagtag |
| zyx1stop (crRNA) | tgactgatggctcgcTTACG |
| Strain | Genotype | Available from |
| N2 | Caenorhabditis elegans | CGC |
| UN1502 | xbls1502[Y66H1Bp::GFP::ACT-1, pRF4(rol-6(su1006))] | Cram Lab |
| UN2013 | zyx-1(xb6) | Cram Lab |
| UN2016 | zyx-1(xb6); xbls1502[Y66H1Bp::GFP::ACT-1, pRF4(rol-6(su1006))] | Cram Lab |
| BC14184 | sEx14184[rCes F42G4.3b::GFP + pCeh361] | CGC |
Acknowledgments
Several strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded through the National Center for Research Resources, National Institutes of Health. The development of the CRISPR allele in this work was a class project of BIO4108 Cell and Developmental Biology Laboratory at Baylor University.
References
Funding
This work is supported by a National Science Foundation/Molecular and Cellular Biosciences–U.S. Israel Binational Science Foundation award (1816640) to E.J.C..
Reviewed By
Shoichiro OnoHistory
Received: September 28, 2021Revision received: October 13, 2021
Accepted: October 15, 2021
Published: October 21, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Castaneda, PG; Wu, N; Qiu, Z; Lee, M; Cram, EJ (2021). ZYX-1/Zyxin plays a minor role in oocyte transit through the spermatheca in C. elegans. microPublication Biology. 10.17912/micropub.biology.000489.Download: RIS BibTeX