Abstract
The heterotrimeric Replication Protein A (RPA) complex preserves genome integrity by protecting the single-stranded DNA that becomes exposed during repair, replication, and recombination. Its two biggest subunits, Rfa1p and Rfa2p (as named in S. cerevisiae) contact DNA and interact with other partners, while the smallest Rfa3p subunit is considered to fulfill a structural role. Perhaps because of this, mostly Rfa1p and eventually Rfa2p are used for microscopy studies upon tagging them with fluorophores. In this work, we explore the behavior of GFP-tagged Rfa3p basally and in response to DNA damage conditions and compare it with tagged Rfa1p. We find that fluorescent Rfa3p yields signals that are (or are detected) significantly more frequent(ly). By making a careful comparison with our own and with previously published data, we propose that Rfa3p, by virtue of its scaffolding role, may reach single-stranded DNA sites first thus serving to nucleate the full RPA complex.
Description
The conserved, heterotrimeric RPA complex is involved in genome stability maintenance at multiple levels. Given its affinity for single-stranded DNA, which it protects from unwanted degradation, this complex is implicated in DNA repair, replication, and recombination (Maréchal and Zou 2015). In Saccharomyces cerevisiae, its three subunits are called Rfa1p, Rfa2p and Rfa3p. The biggest ones, Rfa1p and Rfa2p, contact DNA directly and are also responsible for the interaction with most protein partners (for a comprehensive curation see (Maréchal and Zou 2015)). As such, their fluorophore-tagged versions have recurrently been used in microscopy studies to monitor the implication of the RPA complex in the aforementioned processes ((Lisby et al. 2004; Ivanova et al. 2020; Wong et al. 2020), to cite some examples). Rfa3p is the smallest subunit, with a theoretical molecular weight of 14 kDa. Its position within the 3D-structure (Yates et al. 2018) as well as other studies (Bochkareva et al. 2002) let think that Rfa3p fulfills an exclusively structural role, probably explaining why it has attracted so little interest for microscopy approaches. Only one work, to our knowledge, has simultaneously assessed the subcellular distribution of the C-terminally tagged versions of the three subunits, concluding they all mainly share a nuclear localization (Belanger et al. 2011). Yet, that work did not assess their potentially similar (or different) response to replication challenges or to DNA damage.
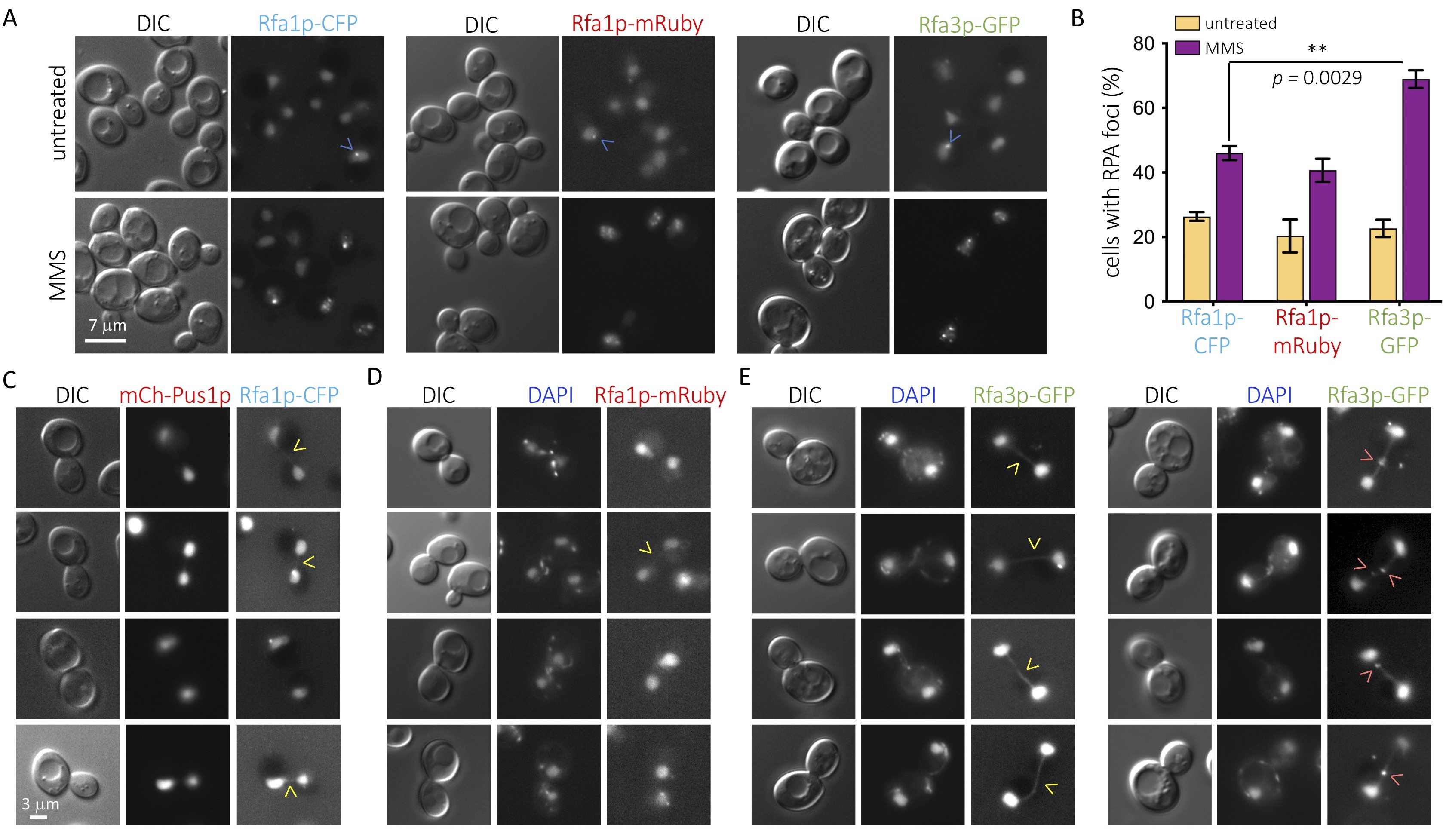
We decided to assess how the C-terminally, fluorophore-tagged versions of Rfa1p and Rfa3p compare to each other under basal conditions and in response to the alkylating agent methyl metanesulfonate (MMS). By damaging nucleotide bases, MMS provokes the accumulation of single-stranded DNA after the passage of the replication fork, thus inducing RPA participation (Wong et al. 2020). Not only was Rfa3p-GFP localization mainly nuclear, as reported (Belanger et al. 2011) but, as tagged Rfa1p, it also congregated spontaneously in the shape of foci under basal conditions (Figure 1A, upper panels, blue arrows), most likely indicative of basal DNA damage or physiological replication-associated single-stranded DNA tracts. Also as fluorescently tagged Rfa1p, which accumulates in post-replicative damaged territories in the shape of foci (Wong et al. 2020), Rfa3p-GFP associated in numerous foci in response to MMS (Figure 1A, bottom panels, blue arrows). Thus, we acknowledge that fluorophore-tagged Rfa3p behaves similarly to its tagged Rfa1p counterpart. Yet, when we quantified the percentage of cells displaying at least one nuclear focus, we observed that the values were reproducibly higher for the Rfa3p-tagged strain in response to MMS (Figure 1B). This was unlikely due to the type of appended fluorophore, since changing Rfa1p tag from CFP to 2 mRuby moieties did not alter the number of detected events, which always remained less abundant than when following Rfa3p (Figure 1B, asterisks), and basal values in the untreated cultures were identical for all three strains. Thus, monitoring of fluorescently tagged Rfa3p could represent a more sensitive means to study the participation, in a challenged DNA scenario, of the RPA complex.
Rfa1p and Rfa2p are also known as markers of the presence of ultra-fine bridges (UFBs) during mitosis (Germann et al. 2014; Liu et al. 2014; Ivanova et al. 2020). These structures mostly result from the failure to segregate unreplicated regions, either naturally difficult-to-replicate sequences or after a stress slowing down replication forks during the preceding synthesis phase. This makes them refractory to DAPI staining, as they lack chromatin features, but they are rich in single stranded DNA portions, thus prone to binding by RPA. UFBs presence in mitoses of unperturbed, cycling cells can be revealed by fluorescent Rfa1p in roughly 30 % of the cases, which manifest as weak, faint signals in the shape of a rope that would connect mother and daughter nuclei (Germann et al. 2014). The same events can be detected in 44 % of mitoses by using fluorescent Rfa2p yet not in the shape of a connecting rope, but of a neat focus somewhere in between both nuclei (Ivanova et al. 2020). We therefore explored whether Rfa3p-GFP could somehow illuminate mitosis-associated events in untreated cultures. As reported, and irrespective of the tag, Rfa1p only modestly marked bridges connecting daughter and mother cells’ nuclei during mitosis (Figure 1C,D). In sharp contrast, almost every mitosis observed in Rfa3p-GFP cells was recognizable by a neat, well-defined rope-like signal connecting both nuclei (Figure 1E, left panel). Further, in roughly half of these cases there was, additionally, one or even two defined Rfa3p foci superimposed on the rope (Figure 1E, right panel). This indicates again that fluorescent Rfa3p provides more sensitive detection than its counterparts Rfa1p or Rfa2p.
Our data have revealed that using fluorescent Rfa3p increases the sensitivity with which single stranded DNA-associated events can be detected, whether during mitosis, or in response to DNA-damaging agents. In a way, the consideration of our and other’s data from unperturbed mitoses suggest that detection sensitivity inversely correlates with the size of the tagged subunit. In this case, our data could imply that the rate of mitosis naturally concurring with at least one UFB has been underestimated. Yet, careful quantification of the time requested prior to cell division by Rfa2p foci-positive cells demonstrated these took longer than Rfa2p foci-negative ones (Ivanova et al. 2020), suggesting that UFBs-possessing cells can be faithfully traced with the Rfa2p-GFP tool. Alternatively, our data could mean that, despite acting as a heterotrimeric complex, the three subunits of RPA do not reach the place of action simultaneously but may assemble sequentially on-site. In this sense, and in agreement with Rfa3p fulfilling a structural role (Bochkareva et al. 2002; Yates et al. 2018), Rfa3p could reach the single stranded DNA region first and then trigger the nucleation of the full complex. This would increase its residence time at those locations, which would translate into an improved detection rate by microscopy approaches. Given the poor characterization of Rfa3p properties in the literature, it will be worth exploring this modular, on-site assembly hypothesis.
Methods
Request a detailed protocolSaccharomyces cerevisiae cells were grown at 25°C in rich YPD medium. All experiments were performed with exponentially growing cells. For microscopy analyses, prior to imaging, cells were incubated for 20 min with 4 µg/mL DAPI to visualize the nuclear DNA. Then, 1 mL of the culture of interest was centrifuged, the supernatant was thrown away and the pellet was resuspended in the remaining 50 μL. Next, 3 μL of this cell suspension was directly mounted on a coverslip for immediate imaging of full cells, by using the Differential Interference Contrast (DIC), and fluorescent signals by using the adequate wavelength. Imaging was achieved using a Zeiss Axioimager Z2 microscope and Metamorph software. Images were acquired at 20–23°C. Subsequent image visualization and analysis were performed with Image J v2.0.0-rc-69/1.52i. The determination of the percentage of cells displaying at least one focus per nucleus was done through visual inspection by the experimenters. GraphPad Prism was used to plot and statistically analyze the results.
Reagents
The RFA1-CFP (mCherry-PUS1::URA3) strain (MM-03) (Lisby et al. 2004), RFA1-2xmRuby::URA3 (MM-170) (Wong et al. 2020) and RFA3-GFP::HIS3 (MM-286) (Huh et al. 2003) are W303 strains corrected for the RAD5 gene.
Acknowledgments
We thank Vincent Géli for the gift of the Rfa1p-CFP strain, Helle Ulrich for the Rfa1p-2xmRuby strain, and Alenka Čopič for the Rfa3p-GFP strain. We acknowledge the imaging facility MRI, a member of the national infrastructure France-BioImaging, supported by the French National Research Agency (ANR-10-INBS-04, Investissements d’avenir).
References
Funding
This research was funded by the ATIP-Avenir program, La Ligue contre le Cancer et l’Institut National du Cancer (PLBIO19-098 INCA_13832), France.
Reviewed By
AnonymousHistory
Received: July 22, 2021Revision received: October 22, 2021
Accepted: October 22, 2021
Published: October 27, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ramonatxo, A; Moriel-Carretero, M (2021). Microscopy analysis of the smallest subunit of the RPA complex, Rfa3p, prompts consideration of how RPA subunits gather at single-stranded DNA sites. microPublication Biology. 10.17912/micropub.biology.000493.Download: RIS BibTeX