Abstract
Calcium-Dependent Protein Kinases (CDPKs) translate calcium ion (Ca2+) signals into direct phosphorylation of proteins involved in stress response and plant growth. To get a clear picture of CDPKs functions, we must identify and explore the CDPKs targets and their respective roles in plant physiology. Here, we present a manually curated Oryza sativa L. CDPK Protein-Protein Interaction Network (CDPK-OsPPIN). The CDPK-OsPPIN provides an interactive graphical tool to assist hypothesis generation by researchers investigating CDPK roles and functional diversity.
Description
Calcium-Dependent Protein Kinases (CDPKs) are essential translators of calcium ion (Ca2+) signaling in protists (Zhang and Choi, 2001), green algae (Valmonte et al., 2014), and plants (Harmon et al., 1987; Roberts, 1993). The ion Ca2+ is a key second messenger of diverse signaling pathways conveying environmental and developmental cues (Dodd et al., 2010). External and internal signals trigger transient changes of cytosolic Ca2+ ([Ca2+]cyt) levels. These changes in [Ca2+]cyt levels can be perceived by Ca2+ binding proteins, such as Calmodulins (CaMs), Calmodulin-Like proteins (CMLs), Calcineurin B-Like proteins (CBLs), and CDPKs (Dodd et al., 2010). The perception of Ca2+ causes conformational changes in CaMs, CBLs, and CDPKs that allow specific protein-protein interactions. After Ca2+-induced conformational changes, CaMs interact with target proteins and recruit Calmodulin-dependent Protein Kinases (CaMKs) to phosphorylate the CaM targets (Zhang and Lu, 2003). Interestingly, CDPKs were proposed to result from a gene fusion between CaM and CaMKs genes (Zhang and Choi, 2001). Consequently, CDPKs are unique in their ability to sense and decode Ca2+ signals by directly phosphorylating specific targets.
CDPKs have a variable N-terminal domain, a serine/threonine kinase domain, and a regulatory calmodulin-like domain (CaM-LD). These kinases also possess an auto-inhibitory junction region that restrains their catalytic activity. The binding of Ca2+ to the EF-hands domains triggers a conformational change that exposes the kinase domain and activates the CDPK. The EF-hands of CDPKs vary in their affinity to Ca2+ (Harmon et al., 2000), suggesting that different CDPKs might respond to different Ca2+ concentrations. Ca2+ signals depend on the stimulus and differ in frequency of oscillation, amplitude, and duration (Dodd et al., 2010). The timing of expression and spatial distribution of CDPKs can also add specificity to the Ca2+ signal decoding (Harmon et al., 2000). However, how CDPKs determine the phosphorylation of their interacting proteins is still largely unknown. For instance, Wang et al., 2011 showed that although both OsCPK2 and OsCPK26 interact with OsCPK25/26-Interacting Protein 30 (OIP30), only OsCPK26 can phosphorylate it.
CDPKs are involved in mediating plant stress responses. In rice (Oryza sativa L.), OsCPK4, OsCPK10, and OsCPK13 are involved in drought stress tolerance (Saijo et al., 2000; Campo et al., 2014; Bundó and Coca, 2017; Wang et al., 2018). OsCPK4 and OsCPK10 enhance blast disease resistance (Bundó and Coca, 2016, 2017), while OsCPK12 and OsCPK18 seem to negatively regulate plant immunity (Asano et al., 2012; Xie et al., 2014). OsCPK12, OsCPK13, and OsCPK21 are involved in salt stress tolerance (Saijo et al., 2000; Asano et al., 2011, 2012), while OsCPK13 and OsCPK17 are involved in cold stress tolerance (Saijo et al., 2000; Almadanim et al., 2017). CDPKs also regulate central metabolism (recently reviewed in Alves et al., 2021), suggesting that they can meditate growth-stress response balance in stressful conditions. However, to fully understand CDPKs’ function, mapping their interactions and phosphorylation targets is necessary.
Rice CDPKs are a large family of 30 members (Asano et al., 2005; Alves et al., 2021) still poorly characterized. Specifically, knowledge of their interactors, which define their function, is still scarce and lacking systematization. For instance, as of October 2021, BioGrid (v.3.5) reported 81,044 physical interactions in Arabidopsis thaliana but only 346 in rice – 236 of which result from a single experiment of affinity capture followed by mass spectrometry analysis (Stark et al., 2006; Biswal et al., 2019). Adding to this, the available information on rice-CDPKs can be challenging to retrieve due to the lack of standardization of gene nomenclature (see Material and Methods section). Working with rice CDPKs for over a decade, our lab felt the need to collect available functional information on CDPK interaction partners, systematizing it to make it readily available to the scientific community. A standard tool to organize and visualize Protein-Protein Interactions (PPI) is PPI Networks (PPIN). PPINs are a graphical representation and integration of large volumes of data and facilitates quick consults and the formulation of new hypotheses.
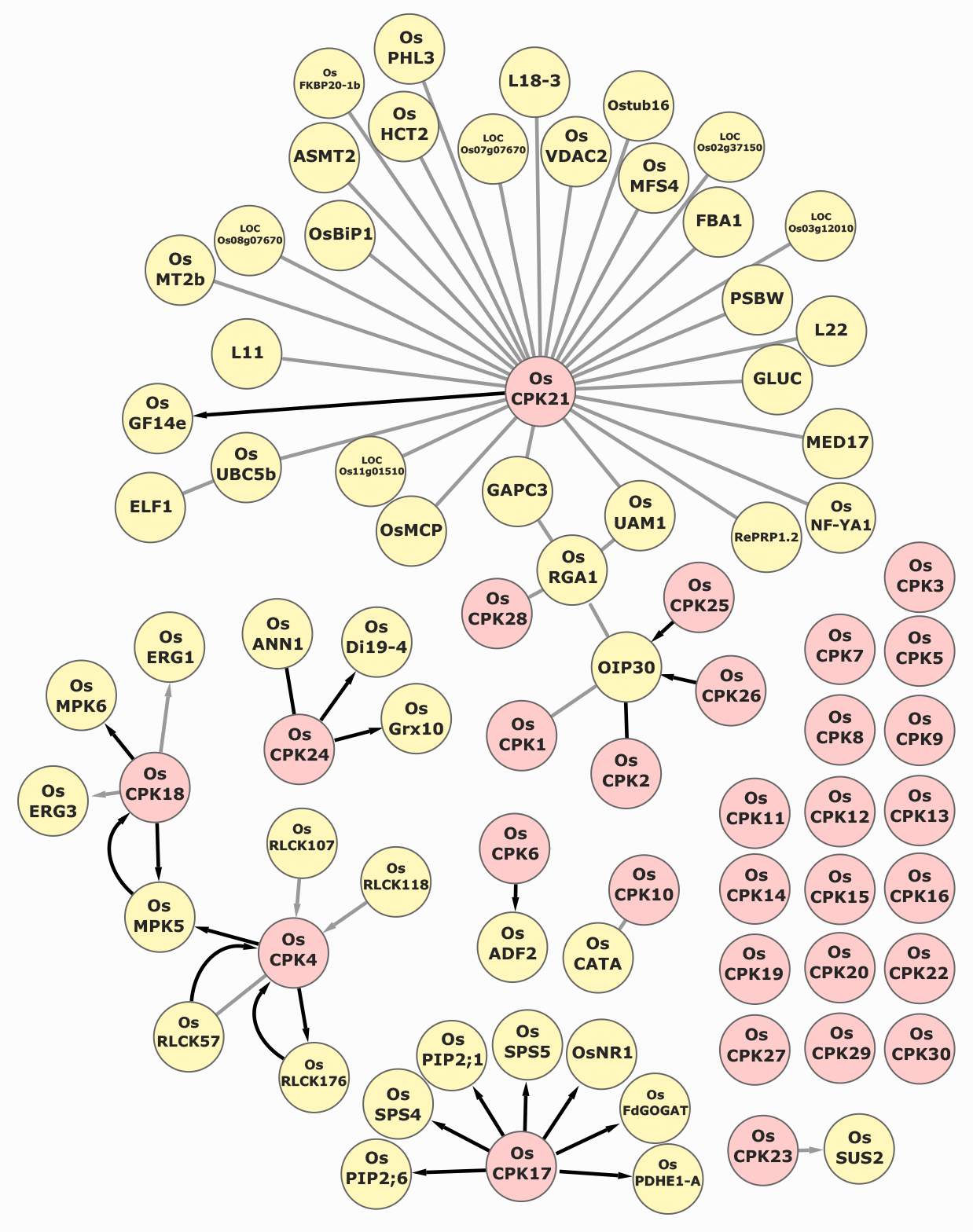
Here, we report the development of an Oryza sativa CDPKs PPIN (CDPK-OsPPIN, Figure 1). The interactive network can be found in (https://bit.ly/3205Tr9). The CDPK-OsPPIN will also be accessible on the web pages of the rice CDPKs of the RAP-DB (https://rapdb.dna.affrc.go.jp) (Kawahara et al., 2013; Sakai et al., 2013). The CDPK-OsPPIN provides a graphical and interactive interface to explore rice CDPK protein interactions and their involvement in rice signaling pathways coordinating plant fitness. To build the CDPK-OsPPIN, we compiled the literature available on the targets of OsCPKs. Additionally, we searched the interaction partners of the CDPK targets in the BioGrid repository (Stark et al., 2006). The CDPK-OsPPIN was built using Cytoscape (v. 3.9.0) and is nested in the Ndex (v2.5.2) platform (https://www.ndexbio.org) (Shannon et al., 2003; Pratt et al., 2015). The Ndex platform allows for a searchable and interactive visualization of the CDPK repository. The nodes (proteins) are clickable and show the following information: Locus ID (MSU and RAP-DB) (Kawahara et al., 2013; Sakai et al., 2013), UniProt ID, other gene/protein symbols, molecular function, biological process, subcellular localization, and Gene Ontology (GO) terms. The interactions between proteins are represented by edges. The edges are also clickable and display the methodology used to determine each interaction and appear between the protein names (e.g., OsCPK21 (Y2H) PSBW) – empirically demonstrated phosphorylation (if tested), the phosphorylated residue (if applicable), and the literature reference that demonstrated the PPI with a clickable DOI. Phosphorylation is indicated by arrows on the edges. The PPIs were demonstrated by: Yeast-Two-Hybrid (Y2H) screenings, Bimolecular Fluorescence Complementation (BiFC), co-immunoprecipitation (Co-IP), in vitro phosphorylation, in vivo phosphorylation, protein identification by affinity capture followed by mass spectrometry, phosphoproteomic, and interaction with target-specific peptides. The number of experiments supporting each interaction is represented by the color of the edge, gray or black, indicating information from one or more independent assays, respectively. This representation allows the user to promptly infer the reliability of specific interactions – most PPIs represented in the network result from more than one low-throughput experiment. The protein and PPI metadata can also be retrieved in the ‘table format’ generated by the platform. The Ndex platform also allows queries that generate sub-networks. For instance, the query ‘nucleus’ will return a sub-network of nuclear proteins and their interactors. So far, CDPK-OsPPIN contains 89 proteins with 62 connections manually curated. The CDPK repository will be updated with manually curated data from future studies.
The CDPK-OsPPIN provides a graphical interface to facilitate hypothesis generation to explore the biological functions, functional divergence, and redundancy of CDPKs.
Methods
Request a detailed protocolThe protein network is available at the Ndex platform (https://bit.ly/3205Tr9). The interaction data for the network were manually curated from the available literature. The protein-protein interaction network was built in Cytoscape (v. 3.9.0) (Shannon et al., 2003). The nodes were displayed in organic layout. The protein annotations such as GO terms, subcellular localization, molecular function, and biological function were obtained from the Rice Genome Annotation Project (Kawahara et al., 2013).
Reagents
We adopted the following nomenclature when referring to a single member of the CDPK family is OsCPKX, where X refers to the number of the CPK. We suggest the adoption of this nomenclature by the scientific community. Here is the full list of members rice CDPK family, with the Rice Annotation Project Database (RAP-DB) Locus ID (Kawahara et al., 2013; Sakai et al., 2013). OsCPK1, Os01g0622600; OsCPK2, Os01g0808400; OsCPK3, Os01g0832300; OsCPK4, Os02g0126400; OsCPK5, Os02g0685900; OsCPK6, Os02g0832000; OsCPK7, Os03g0128700; OsCPK8, Os03g0808600; OsCPK9, Os03g0688300; OsCPK10, Os03g0788500 ; OsCPK11, Os03g0789000; OsCPK12, Os04g0560600; OsCPK13, Os04g0584600; OsCPK14, Os05g0491900; OsCPK15, Os05g0585500; OsCPK16, Os05g0467000; OsCPK17, Os07g0161600; OsCPK18, Os07g0409900; OsCPK19, Os07g0515100; OsCPK20, Os07g0568600; OsCPK21, Os08g0540400; OsCPK22, Os09g0514200; OsCPK23, Os10g0539600; OsCPK24, Os11g0171500; OsCPK25, Os11g0136600; OsCPK26, Os12g0133500; OsCPK27, Os12g0486600; OsCPK28, Os12g0169800; OsCPK29, Os12g0230200; OsCPK30, Os07g0641200.
Acknowledgments
We would like to thank Hugo Cordeiro (ITQB-NOVA) and Luís Morgado (ITQB-NOVA) for the help in making the CDPK-OsPPIN online at PRPlants web-page.
References
Funding
We acknowledge the Portuguese Fundação para a Ciência e a Tecnologia (FCT) for a fellowship for JM (PD/BD/06917/2020) and for a contract for CCM (PTDC/BIA-FBT/31211/2017). Our work was supported by PTDC/BIA-FBT/31211/2017 and by the FCT research fund GREEN-it ‘Bioresources4sustainability’ (UIDB/04551/2020). The funding sources had no involvement in analyses, interpretation of data, writing, or in the decision to submit this paper.
Reviewed By
AnonymousHistory
Received: November 25, 2021Revision received: December 28, 2021
Accepted: January 12, 2022
Published: January 26, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Marques, J; Matiolli, CC; Abreu, IA (2022). Visualization of a curated Oryza sativa L. CDPKs Protein-Protein Interaction Network (CDPK-OsPPIN). microPublication Biology. 10.17912/micropub.biology.000513.Download: RIS BibTeX