Abstract
While evaluating the effect on lifespan of decreased ribosomal protein (Rp) expression in Drosophila, we discovered a potential function in the same process for the Molybdenum cofactor synthesis 1 (Mocs1) gene. We utilized the UAS-GAL4 inducible system, by crossing tissue-specific GAL4 drivers to the Harvard Drosophila Transgenic RNAi Project (TrIP) responder lines for Rp gene knockdown. We also employed a negative control that knocked down a gene unrelated to Drosophila (GAL4). Relative to the genetic background in which no driven transgenes were present, lifespan was significantly lengthened in females, both for Rp knockdown and the negative GAL4 control. We reasoned that the Mocs1 gene, located immediately downstream of the integration site on the third chromosome where all the TrIP responders are targeted might be responsible for the lifespan effects observed, due to the potential for upregulation using the UAS-GAL4 system. We repeated the lifespan experiment using an enhancer trap in the same location as the TrIP transgenes, and found that lifespan was significantly lengthened in females that possessed both the driver and responder, relative to controls, implicating Mocs1 in the biology of aging.
Description
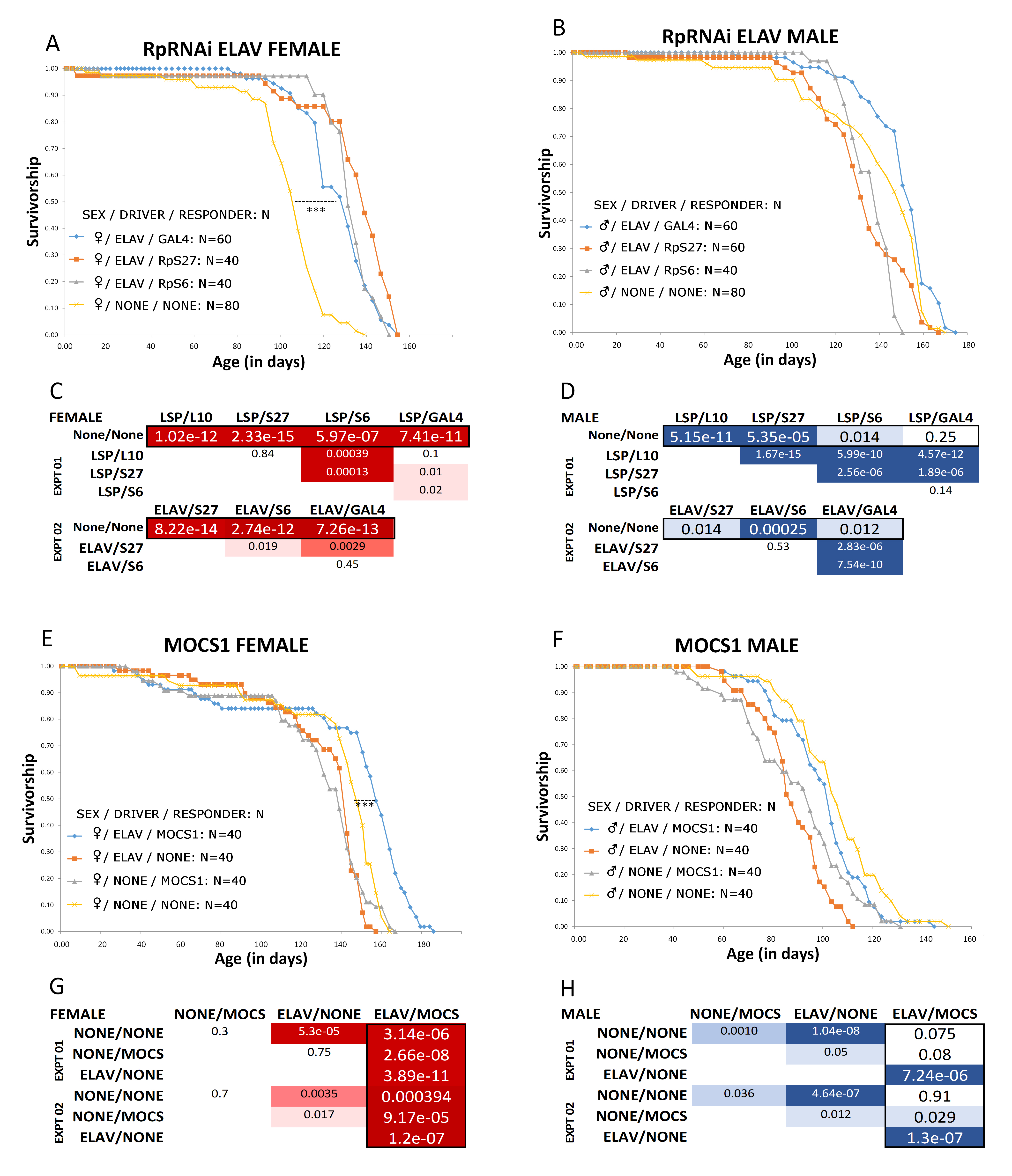
The experiment described in this paper resulted from a failed negative control in a study examining the effect of reducing ribosomal protein (Rp) gene expression on lifespan in Drosophila. Reduction in several different Rps has been shown to lengthen lifespan in a variety of model organisms (C. elegans, S. cerevisiae, D. melanogaster among others Steffen et al. 2008, Bell et al. 2009, Lindquist et al. 2011) presumably via impacts on Target of Rapamycin (TOR) signaling and possibly also mitochondrial function (Riera et al. 2016). As a means of knocking down Rp gene expression in vivo, we made use of the modularized miss-expression system consisting of GAL4 drivers (genetic strains of flies that express the yeast GAL4 transcription factor tissue-specifically) and responders (genetic strains of flies that possess GAL4 inducible transgenes expressing a gene of interest) (Rørth et al. 1998). When drivers and responders are crossed to each other, GAL4 induction of the gene of interest can be observed in the progeny. For the Rp experiment, we specifically employed transgenic RNAi responder lines from the Harvard Drosophila Transgenic RNAi Project (TrIP) where inducible transgenes expressing dsRNA against specific Rps were all integrated into a targeted locus on the third chromosome. This locus had been selected based on extensive expression analysis designed to minimize position effects that might shut down a transgene due to genomic location (Zirin et al. 2020). All the transgenes we used were located ~40 bp upstream of a gene called Mocs1 (CG33048), which encodes a cofactor required by enzymes that utilize Molybdenum. In our RpRNAi lifespan experiment, we used maternally inherited neuronal and fat body GAL4 inducers (drivers) to knock down Rp gene expression (paternally inherited RNAi responders) in these specific (neuronal and fat body) tissues where the intersection between nutrient sensing and metabolism correlates with lifespan modulation (Shen et al. 2009, Hoffman et al. 2013, Fabian et al. 2021). The direction of the cross appeared to matter (i.e., which parent passed the driver or responder to the experimental offspring) as results were equivocal for the reciprocal cross in a pilot. As a first negative control for the RpRNAi experiment, we used progeny from a cross between the original strain the TrIP project used to target the RNAi transgenes (this contains the att-P2 “docking site” but no inducible GAL4 transgene) and the w[1118] isogenic strain which represented the driver background (see reagents). This combination served as a non-isogenic background control since the TrIP lines themselves could not be isogenized efficiently. (These lines are in a genetic background that makes it very difficult to follow the presence or absence of the transgenes by eye through multiple generations, necessitating a molecular approach that would effectively double the length of time in which to complete an already lengthy experiment – see methods below.) As a second negative control for the RpRNAi experiment, we used a TrIP line with a GAL4 inducible transgene expressing RNAi against GAL4 itself (this is a recommended control line from the TrIP project, see reagents; Zirin et al. 2020).
The surprising result was that all lines induced by GAL4, including the negative control driven GAL4 RNAi responder, showed a statistically significant lifespan extension in females relative to the non-induced genetic background control (Figure 1A, B, C and D). This experiment was repeated five times, with concealed genotypes using dLife software (Linford et al. 2013) to ensure data collection was unbiased and blind. A search of the literature regarding the effects of GAL4 induction and/or RNAi in Drosophila on lifespan were inconclusive, and then, only, for ubiquitous (as opposed to tissue-specific) drivers (Alic et al. 2012; Slade and Staveley 2015). Nevertheless, this UAS-GAL4 modularized system has been widely used for lifespan studies (Chavrous et al. 2001, Kapahi et al. 2004, Ruzzi et al. 2020). In addition, there is evidence for a QTL in the vicinity of the Mocs1 locus that correlates with a longer lifespan in Drosophila (Tahoe et al. 2002 Mocs1 called low xanthine dehydrogenase (lxd) at the time). We reasoned that driving expression in the Mocs1 region might lead to its upregulation, which in turn may lengthen lifespan based on a hypothetical role for this gene’s product in regulating cellular protection in redox biochemistry (Zhang and Gladyshev, 2008). Thus, we repeated the lifespan experiment, this time using an enhancer trap (an inducible GAL4 transgene that will drive expression of genes near which it is located) in the same location (and correct orientation with respect to Mocs1) as the targeted RNAi transgenes from the TrIP resource (see reagents). Additionally, we isogenized the genetic background of both a single driver (ELAV) and the responder (enhancer trap) for ten generations, a critical procedure that controls for genetic background, which, as mentioned, is not feasible using the genotypes from the Harvard TrIP resource. We chose to focus on the ELAV pan-neuronal driver because Mocs1 appears to be expressed predominantly in the nervous system (Schauer et al. 2013, Brown et al. 2014). The data are shown in Figure 1: E, F, G, and H. While the inferred position effect induction of MocsI was not directly measured by RTQPCR (see proposed future work below), we did observe a statistically significant lifespan extension in the experimental group (Drosophila bearing both the MOCS1 enhancer trap and the ELAV driver), albeit, and again, only in females.
How would putative upregulation of Mocs1 contribute to lifespan extension? Molybdenum is a transition metal utilized across prokaryote and eukaryote taxa in metabolism, but it typically requires an organic compound (a pyranopterin) for enzyme catalytic functionality (Mendel, 2013). Mocs1 in humans encodes two protein products via a complex alternative splicing process: a MOCS1A protein and a MOCS1AB fusion protein, both of which catalyze the first step (converting GTP (Guanosine triphosphate) into cPMP (cyclic pyranopterin monophosphate) in a complex pathway that produces the pyranopterin cofactor (Molybdenum Cofactor or “Moco”, Leimkühler, 2017). It is not clear whether the same complex alternative splicing process occurs in Drosophila, however the Mocs1 gene structure is largely conserved (Gray and Nichols, 2000). Several critical redox enzymes use Moco, performing essential physiological and environmental functions, involving the nitrogen, sulphur and carbon cycles (Zhang and Gladyshev, 2008, Marelja et al. 2018). In flies, enzymes in the MOCO synthesis pathway and enzymes that require MOCO cause eye colour phenotypes that have provided early models for physiological biochemistry (Marelja et al. 2018). In humans, mutations in the Moco synthesis pathway segregate with severe disease (early childhood lethal) primarily resulting from sulfite oxidase deficiency, required for cysteine catabolism (Schwarz, 2016). Sulfite oxidase is located in the intermembrane space of the mitochondrion, where electrons resulting from cysteine oxidation are passed to acceptors in the electron transport chain (Hille et al. 2014). The Drosophila homolog of sulfite oxidase (“shopper”) is required in glial cells, and modulates glutamate metabolism, required for normal neuronal excitation, with loss of function alleles displaying locomotory and behavioral defects (Otto et al. 2018). Moco-requiring enzymes also have “moonlighting” roles in additional biochemical processes including functions related to cellular protection and mitochondrial respiration (Gladwin et al. 2005). The Mocs1 gene product is therefore feasibly situated to contribute to lifespan modulation, given the established intersection between mitochondrial homeostasis and pathways known to influence lifespan (Target of Rapamycin (TOR) inhibition, Insulin and insulin-like pathway signaling (IIS), Caloric or Dietary restriction; (Kapahi et al. 2004, Skorupa et al. 2008, Slack et al. 2011).
What explains the sex-specific results for both RpRNAi and MOCS1 experiments? A recent study provides suggestive evidence that loss of function in the Drosophila Mocs1 gene may play a role in regulating male aggression (Ramin et al. 2019), which may affect lifespan in segregated males (as per our experimental design). There are also sex-specific responses in flies to dietary Molybdate, where treatment with low concentrations enhanced antioxidant activity whereas high concentrations were detrimental, and males were more sensitive to these effects than females (Perkhulyn et al. 2017). Given that aging involves a balance between reproduction vs. somatic maintenance, and the unique and costly metabolic requirements for making eggs, it is perhaps not surprising that sex-specific differences are often observed in both stress resistance (females having higher antioxidant potential) and aging (Tower 2015, Perkhulyn et al. 2017). Also, mitochondria are maternally inherited, thus more likely to be optimized in females for lifespan and stress resistance by natural selection (sexual antagonism Tower 2006). Related to sex-specificity, our observation that lifespan extension was only observed (in females) when the drivers were maternally inherited suggests a maternal effect, supported by similar published results concerning the UAS-GAL4 system (Slade and Slaveley 2015). One culprit may be the arthropod bacterial parasite Wolbachia which has been shown to enhance tolerance to iron stress, which may have a knock-on effect in mitochondrial turnover (Kosmidis et al. 2014).
Future work to solidify Mocs1 involvement in lifespan extension would include quantitative expression analysis (RNAseq, RTQPCR, etc.) to confirm upregulation of Mocs1 in the context of lifespan extension, and interaction studies to fit Moco biosynthesis into established regulatory networks that contribute to the biology of aging. PCR and antibiotic treatments might resolve any Wolbachia involvement. Finally, this study provides cautionary evidence when using the Harvard third chromosome TrIP lines in any experimental analysis of aging in Drosophila.
Methods
Request a detailed protocolFor the MOCS1 experiment, isogenization was performed by ten generations of back crosses to an isogenized white eyed-line (w1118 BL5905 see Reagents below) by following the red eye color reporter (w+) located on both the driver and responder transgenes. The isogenized driver was tested for functionality by crossing to a GFP responder and examining the progeny by fluorescence microscopy. The presence of the responder transgene in the isogenized stock was tested for by PCR. (Note isogenizing was not possible for the RpRNAi TrIP lines because the transgene reporters were not easily distinguishable by eye). Lifespans were measured using established protocols (Linford et al. 2013). Typically, 2-3 replicate vials (approximately 40-60 flies of a given sex) were established for each driver/responder combination. A standard cornmeal-yeast-agar food recipe was employed, albeit with rather low sucrose as is habitual in our laboratory (1% where 5-10% is more usual – this means the flies are somewhat calorically restricted (CR), so any lifespan extension is therefore likely additive to CR rather than epistatic). Flies were first collected from uncrowded bottle conditions. Newly eclosed males and females of each relevant genotype were allowed to mate for 48 hours before lifespan data were collected. Flies were transferred to fresh media every 1-2 days, at which time dead flies were removed and recorded using the dLife system developed in the Pletcher Laboratory (Linford et al. 2013). Constant temperature (22-25°C as measured daily in the lab) and humidity (60% approximate, based on facilities management, no major swings) conditions were maintained, with a 12:12 hour light:dark cycle. Lifespan comparisons between different genotype survivorship curves were carried out using the statistical package R within dLife (Linford et al. 2013). P-values were obtained using the log-rank test.
Reagents
| Fly strain name | Genotype | Description | Bloomington Drosophila Stock center # | Reference |
| ELAV GAL4 Driver | w[*]; P{w[+mC]=GAL4-elav.L}CG16779[3] | Pan-neuronal driver | 8760 | Sink et al. 2001 |
| LSP2 GAL4 Driver | y[1] w[1118]; P{w[+mC]=Lsp2-GAL4.H}3 | Fat body driver | 6357 | Cherbas et al. 2003 |
| RpS6 RNAi Responder | y[1] sc[*] v[1] sev[21]; P{y[+t7.7]v[+t1.8]=TRiP.HMS00413}attP2 | GAL4 inducible responder cloned upstream of the Mocs1 locus; targets RpS6 with RNAi | 32418 | Zirin et al. 2020 |
| RpS27 RNAi Responder | y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01581}attP2 | GAL4 inducible responder cloned upstream of the Mocs1 locus; targets RpS27 with RNAi | 36692 | Zirin et al. 2020 |
| RpL10 RNAi Responder | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02520}attP2 | GAL4 inducible responder cloned upstream of the Mocs1 locus; targets RpL10 with RNAi (only used with LSP2 driver) | 29356 | Zirin et al. 2020 |
| TrIP line background | y[1] v[1]; P{y[+t7.7]=CaryP}attP2 | Att-P2 “landing site” background for TrIP lines | 36303 | Zirin et al. 2020 |
| TrIP RNAi negative control line | y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=VALIUM20-GAL4.1}attP2 | GAL4 inducible responder cloned upstream of the Mocs1 locus; targets GAL4 | 35784 | Zirin et al. 2020 |
| Isogenic control | w[1118] | w (white eye mutant) line isogenic for chromosomes 1,2 and 3 | 5905 | No publication, Flybase ref: RRID:BDSC_5905 |
| Mocs1 enhancer trap | y[1] w[67c23]; P{y[+mDint2] w[+mC]=EPgy2}EY00759 | GAL4 inducible enhancer trap in same location as TrIP landing site (stock # 36303) upstream of Mocs1 | 19808 | Bellen et al. 2004 |
Acknowledgments
We thank Scott Pletcher for his assistance in setting up and running dLife in our lab. We also thank the Kaeberlein lab for their advice and feedback throughout the RpRNAi project.
References
Funding
Funding was provided by a Professional Development Award from the Office of Research & Sponsored Programs at Western Washington University.
Reviewed By
AnonymousHistory
Received: September 23, 2021Revision received: January 7, 2022
Accepted: January 18, 2022
Published: January 25, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Lamont, EI; Lee, M; Burgdorf, D; Ibsen, C; McQualter, J; Sarhan, R; Thompson, O; Schulze, SR (2022). Mocs1 (Molybdenum cofactor synthesis 1) may contribute to lifespan extension in Drosophila. microPublication Biology. 10.17912/micropub.biology.000517.Download: RIS BibTeX