Bharat Chattoo Genome Research Centre, Department of Microbiology & Biotechnology Centre, Faculty of Science, The Maharaja Sayajirao University of Baroda, Gujarat, India
Abstract
Evolutionarily conserved nucleosome assembly protein Nap1 is involved in multiple cellular processes in eukaryotes. In this study, we wanted to explore the role of Nap1 in the life cycle of rice blast fungus Magnaporthe oryzae. The null mutant of M. oryzae NAP1 is viable. However, deletion of NAP1 leads to defects in growth, appressorium morphology, and appressorium turgidity. In the future, plant infection studies can be undertaken to find if these defects lead to compromised virulence of this economically important fungal pathogen.
Description
Rice blast, caused by Magnaporthe oryzae, is a devastating plant disease with epidemics leading to losses of up to 30% of rice harvest yield (Wilson and Talbot 2009). This organism tops the list of ten major fungal pathogens in molecular plant pathology, owing to concerns over food security and economy (Dean et al. 2012). M. oryzae has a unique infection cycle. Infection is initiated when a fungal spore or a conidium attaches itself to the rice leaf cuticle and on receiving the suitable environmental cues, germinates to give rise to a single polarised germ tube. The germ tube later differentiates into a dome-shaped infection structure called the appressorium. Immense turgor pressure is generated within the appressorium which leads to mechanical disruption of the underlying leaf cell wall that facilitates the fungus to invade the plant cell where it proliferates and spreads in the plant tissue. In high humidity, the fungus sporulates out of the host and the spores are spread in the environment via rain splash and wind (Wilson and Talbot 2009). Elucidation of each of these steps is owed to results of extensive research in the last few decades which led to identification of several proteins and pathways involved in the infection cycle. This study was undertaken with the aim to understand the role of a core protein, nucleosome assembly protein (Nap1) in the life cycle of the blast fungus M. oryzae.
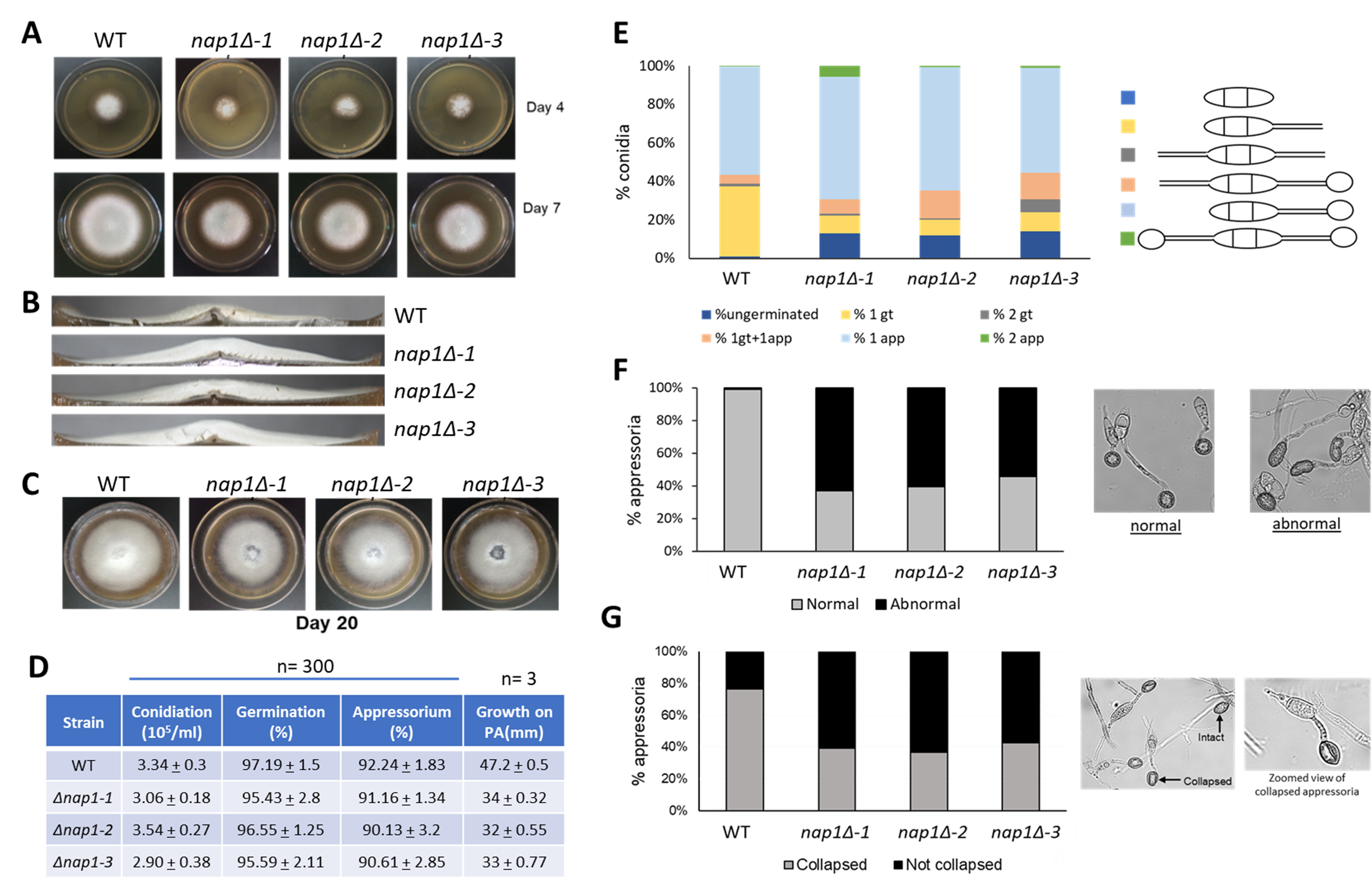
NAP proteins, as the name suggests, are involved in incorporation of histones during nucleosome assembly. They are highly conserved proteins present across all eukaryotes. Nap1 is involved in nucleosome remodelling, cell cycle regulation, transcriptional regulation, and shuttling of histones between the nucleus and cytoplasm (Zlatanova et al. 2007). M. oryzae NAP1 (MGG_06024; NAP1) is a 1549 bp ORF with four introns. The spliced product (1215 bp) gives rise to a 404-amino acid polypeptide that has 46% identity with Saccharomyces cerevisiae Nap1. Using the gene replacement strategy, three independent nap1Δ mutants (nap1Δ-1, nap1Δ-2, nap1Δ-3) were generated and studied. Deletion of this gene was found to be nonessential and was observed to be associated with growth and appressorium formation. The NAP1 null cells show defects in lateral spreading of the colony as observed by the smaller colony size (Fig. 1A). This may also indicate slower growth rate of these mutants. In addition, the mutant colonies appear fluffier with almost double the vertical size of the aerial hyphae mass than the wild-type (Fig. 1B). Colonies of NAP1 null cells also underwent early senescence as seen by autolysis of the older cells at the middle of the colony in the plate (Fig. 1C), indicating that Nap1 may have a role in the aging process. The rate of sporulation in nap1Δ strains is similar to wild-type and nap1Δ conidia germinate and form appressoria within the same time as wild-type (Fig. 1D). However, defective germination was observed in a small percentage of mutant conidia. The number of conidia germinating with two germ tubes was more in the mutants, along with a few germ tubes with two appressoria (Fig. 1E). In addition, the appressorial morphology of mutants was distorted. While appressoria are usually circular or dome-shaped, about 50-60% appressoria in mutants were found to be elongated and abnormal (Fig. 1F). In yeast, Nap1 plays a role in regulating transition between polarized and isotropic growth (Kellogg and Murray 1995). While the polarized growth of the M. oryzae nap1Δ germ tube does transition to isotropic growth of the appressoria, it is not a normal transition. In the infection cycle of M. oryzae, the formation of the appressorium is highly critical for a successful infection. After the spore germinates to form an appressorium, solutes like glycerol and lipids accumulate and generate turgor pressure inside the appressorium. As much as 8 MPa of pressure is generated and with this sheer mechanical pressure, the appressorium breaches the plant cuticle and cell wall, and the fungus penetrates the host tissue (de Jong et al. 1997). To study if Nap1 is involved in generation of appressorial turgor, a cytorrhysis assay was performed (Galhano et al. 2017). An indirect estimate of appressorium turgor pressure is to measure the collapse of the appressorium on addition of exogenous glycerol. In this assay, the appressoria formed by NAP1 null cells showed greater stability in the concentration of glycerol used, while 70-80% of wild-type appressoria collapsed (Fig.1G). This indicates that Nap1 may be involved in regulating appressorial turgidity. From the above results, we can conclude that Nap1 plays an important role in formation of mature dome-shaped appressoria with an optimum turgor pressure.
For future, it will be important to determine whether nap1Δ conidia (with abnormal appressoria) are able to successfully penetrate the leaf tissue and cause disease.
Methods
Request a detailed protocol- Strains and growth conditions:
M. oryzae wild-type (WT) B157 strain (MTCC accession no. 12236; Kachroo et al., 1994) belonging to the international race IC9 was used in this study. M. oryzae B157 was grown on Prune agar (PA) or complete medium (CM) as described previously (Soundararajan et al.. 2004) at 28°C for 8-10 days. The colonies were cultivated on PA medium for 4 days in dark, followed by 4-5 days growth in constant illumination at room temperature. Vegetative growth was assessed by visual observation of the colony morphology and by measuring the colony diameter. Conidia were harvested as described previously (Patkar et al., 2010), followed by microscopic observation of the conidial morphology. Harvested conidia were counted using a hemocytometer. For growth in liquid medium, small pieces of 8 day-old colony were used to inoculate in 30 mL CM at 28°C for 2-3 days. The biomass was collected by filtering through Miracloth (Calbiochem, Darmstadt, Germany) and drying using paper towels. The biomass was frozen and used for DNA extraction following standard protocols (Dellaporta et al.. 1983). Individual isolates were stored as filter stocks at -20°C.
- Gene replacement:
The sequence of Nap1 (MGG_06924) was obtained from M. oryzae genome assembly (Broad Institute, MIT). The knockout cassette was made by fusing three products – Nap1 upstream (Up), marker (Zeocin) ORF, and Nap1 downstream (Dn) in a molar ratio of 1:3:1 (Up:Zeo:Dn) by overlap PCR. The fused product was then amplified using XT-polymerase, precipitated and 2 µg of this amplified DNA was used for protoplast transformation of M. oryzae. Transformants were screened by negative PCR using Nap1 ORF primers and estimating band size differences using cassette primers and nested primers combinations.
- Protoplast transformation:
M. oryzae B157 was inoculated in 30 mL of CM broth and grown for 3 days. The biomass was then filtered with Miracloth, washed with sterile water and the mycelia were resuspended in 30 mL of 1M sorbitol containing 30 mg (1 mg/mL) Trichoderma viridae lytic enzymes (Sigma, St. Louis, USA). This was incubated at 28°C at 100 rpm for 12-16 hours for protoplasting. Next day, the protoplasts were filtered, washed and resuspended in 10 mL of 1 M sorbitol at 4000 rpm for 5 min at 4°C twice. The protoplasts were resuspended in 10 mL STC buffer (1 M sorbitol, 50 mM Tris-HCl pH 8, 50 mM CaCl2.2H2O), washed, and resuspended again in 1 mL STC buffer. The protoplasts were counted using a hemocytometer and a density of 108 cells/mL was maintained. For every transformation, 200 µL of protoplasts was mixed with 2-5 µg of DNA (dissolved in TE buffer) and incubated on ice for 15 min. To this 1 mL of PTC buffer (40% PEG 3350 + STC) was added and incubated at 28°C for 30 min. The whole mixture was transferred to a 15 mL tube containing 3 mL of YEGS (0.2% yeast extract, 1% dextrose, 1 M sorbitol) and was incubated at 28°C at 100 rpm for 12-14 hours. After incubation, 10 mL of molten regeneration medium (0.2% yeast extract, 1% dextrose, 0.4% agarose) was added, mixed well and poured on YEG agar containing 300 µg/mL Zeocin. Appearance of colonies was monitored within 2-3 days of incubation at 28°C. Water was used instead of DNA as negative control.
- Appressorium assay:
Fungal conidia were harvested by gently scraping off the biomass grown on PA plates using a sterile disposable plastic loop. The biomass was then filtered through two-layers of Miracloth. Spores were collected by centrifugation at 13,000 rpm for 4 min and re-suspended in water. Spore concentration of 105 spores/mL was used and 10 µL of spore suspension was placed on hydrophobic cover-slip. This system was incubated in dark in a humid chamber for 12 hours, after which at least 100 conidia were imaged.
- Cytorrhysis assay:
Briefly, 105 spores were allowed to form appressoria for 18 h on coverslips after which water from the conidial sample was removed carefully by pipetting and replaced with 3M glycerol. After 10 minutes in glycerol, ~300 appressoria were imaged and analyzed.
All experiments were performed with three independent deletion mutants and at least twice with reproducible results.
Acknowledgments
We would like to acknowledge late B. B. Chattoo for mentoring SP to initiate the project in his lab, for providing infrastructural support and for his insightful suggestions during the initial part of this project. Bharat Chattoo Genome Research Centre is acknowledged for providing the fungal strain B157 and for initial help with protocols. We thank Rajesh Patkar for helpful suggestions for culturing the fungus and extracting spores. We are also thankful to the members of Bharat Chattoo lab and Molecular Mycology lab for useful discussions.
References
Funding
SP acknowledges SERB-NPDF fellowship (PDF/2015/001000) for the funding. KS acknowledges Department of Biotechnology (DBT) grant in Life Science Research, Education and Training at JNCASR (BT/INF/22/SP27679/2018).
Reviewed By
Douglas KelloggHistory
Received: September 27, 2021Accepted: January 18, 2022
Published: January 24, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Panchal, S; Sanyal, K (2022). Loss of nucleosome assembly protein 1 affects growth and appressorium structure in blast fungus Magnaporthe oryzae. microPublication Biology. 10.17912/micropub.biology.000520.Download: RIS BibTeX