Abstract
Long intergenic non-coding RNAs (lincRNAs) are transcripts longer than 200 nucleotides which are transcribed from regions that do not overlap with protein coding sequences. Reproductive organs express high levels of lincRNAs, yet removal of many lincRNA genes with high and dynamic germline expression did not lead to fertility defects. It was previously suggested this stems from redundant roles of different lincRNA genes. We previously reported engineering C. elegans strains in which we deleted lincRNA genes with high and dynamic expression in the gonad. The individual mutations did not lead to major effects on fertility. Two of those lincRNA genes, linc-9 and linc-20, are highly homologous, suggesting they could perform redundant roles. Here we report that in the double mutant linc-9; linc-20 the brood size and embryonic lethality do not significantly differ from wild-type worms. This could be explained by either lack of fertility roles, or redundancy with other lincRNA genes.
Description
Long non-coding RNAs (lncRNAs) are transcripts which are longer than 200 nucleotides which are not translated into proteins. These transcripts are usually transcribed by RNA Polymerase II, often capped, polyadenylated, and spliced (reviewed in (Blythe et al., 2016; Deniz and Erman, 2017; Fatica and Bozzoni, 2014; Fico et al., 2019; Kopp and Mendell, 2018; Marques and Ponting, 2014; Melissari and Grote, 2016; Shields et al., 2019; Ulitsky and Bartel, 2013)). Long intergenic non-coding RNAs (lincRNAs) are a subclass of lncRNAs which are transcribed from genomic regions which do not code for proteins (Deniz and Erman, 2017). Many lincRNAs are transcribed during gametogenesis, and mammalian testis is the tissue with the highest number of dynamically expressed long non-coding RNAs (Soumillon et al., 2013; Washietl et al., 2014). In many metazoans reverse genetics analysis indicated that knockout or knockdown of lncRNAs which are dynamically expressed in reproductive organs did not lead to loss of fertility (Dai et al., 2019; Ganesh et al., 2020; Goudarzi et al., 2019; Li et al., 2020; Wei et al., 2019 Ishtayeh, 2021 #8555; Zhang et al., 2012; Zhou et al., 2021; Zhu et al., 2020). Several reports suggested this is due to redundancy in the action of two or more lncRNA genes (Goudarzi et al., 2019; Ishtayeh et al., 2021; Wichman et al., 2017). To the best of our knowledge testing this hypothesis has not been published.
We previously reported the analysis of expression of lincRNA genes in the gonad of C. elegans, and the identification of six genes with high and dynamic expression (Ishtayeh et al., 2021). We created strains with complete deletion of these genes using CRISPR. Compared to localized mutation within the gene, complete deletion ensures that no leftover transcript still maintains residual roles, and that transcriptional compensation is not triggered (El-Brolosy et al., 2019; Rossi et al., 2015; Serobyan et al., 2020), thus reducing the possibility for misinterpretation of the roles of the mutated gene. We reported that we did not find any significant reduction in brood size in any of the six lincRNA deletion strains (Ishtayeh et al., 2021). We also did not find any change in several meiotic processes we examined. In that report we noted that two of these lincRNAs, namely linc-9 and linc-20, are highly homologous (over 90% identity) (Ishtayeh et al., 2021). This raises the hypothesis that these lincRNAs genes have similar or redundant roles, and so when only one is deleted, no fertility effect can be detected.
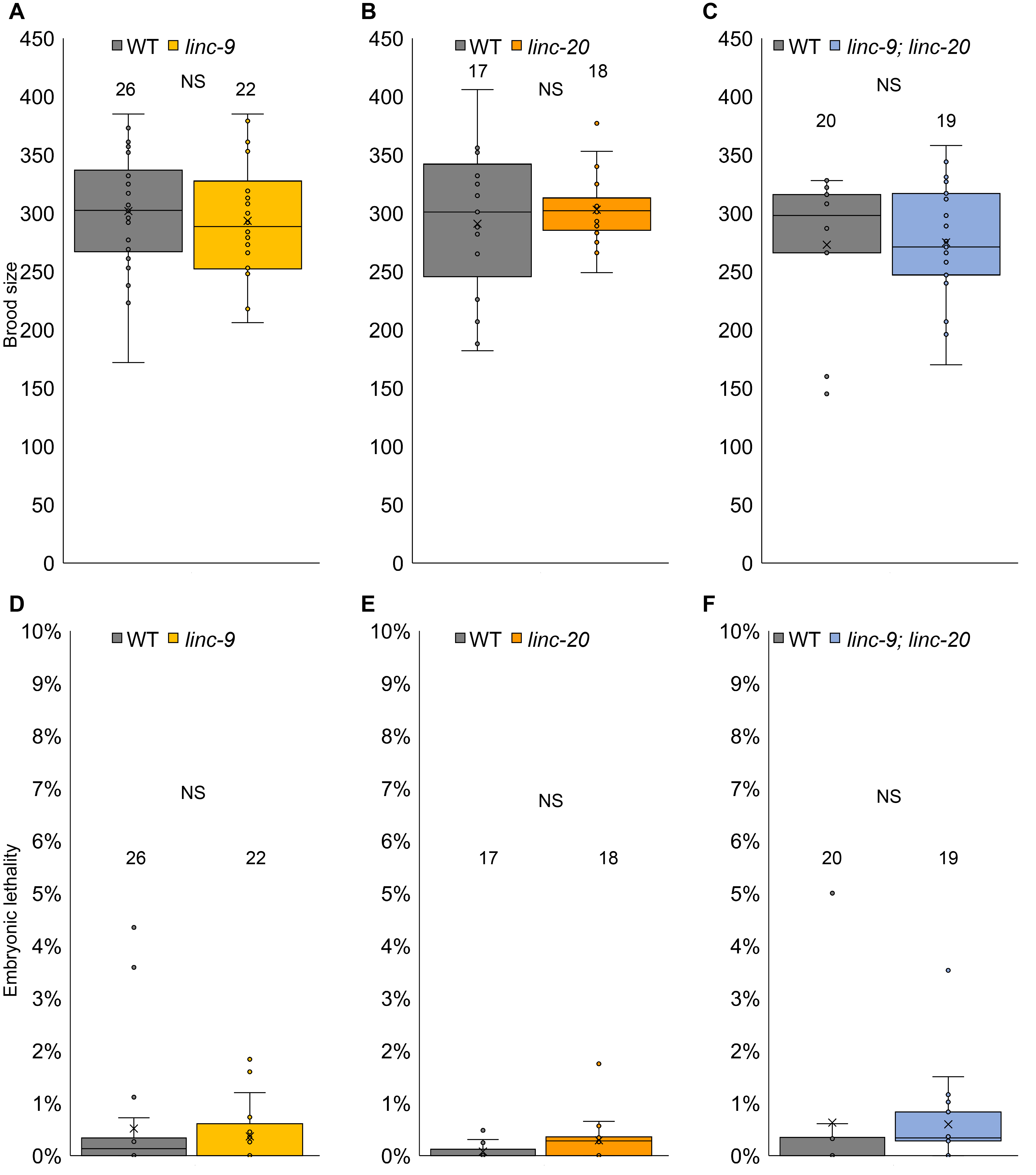
To find if linc-9 and linc-20 have redundant roles in hermaphrodite’s fertility, we crossed the five times outcrossed individual deletion strains to create a linc-9; linc-20 homozygous strain. To compare the fertility of this strain to wild-type worms, we singled L4 larvae on NGM plates, and transferred them to new plates every 24 hours for three days (see methods). We found no significant change in the brood size of the worms between wild type and linc-9; linc-20 worms (Fig. 1, 273±14 and 275±12, n = 20,19 respectively, p value = 0.8 by the two tailed Mann-Whitney test). linc-9; linc-20 brood was similar to those of the single mutants (compare Fig. 1 C to Fig. 1 A, B, and (Ishtayeh et al., 2021)). This result suggests that just like the single mutations, the double mutation of linc-9 and linc-20 does not reduce C. elegans fertility under normal laboratory conditions.
Embryonic lethality (Emb phenotype) was used previously to identify meiotic aberrations (as well as other developmental defects). Neither linc-9 nor linc-20 led to increased embryonic lethality compared to wild-type worms (Ishtayeh et al., 2021). To test if the combined linc-9; linc-20 leads to increased embryonic lethality, we scored the percentage of unhatched embryos laid by wild-type vs. linc-9; linc-20 worms. We found no significant change between the strains (Fig. 1B, 0.6%±0.3% and 0.6±%0.2%, n= 20,19 respectively, p value = 0.4 by the two tailed Mann-Whitney test). linc-9; linc-20 embryonic lethality was similar to those of the single mutants (compare Fig. 1 F to Fig. 1 D, E, and (Ishtayeh et al., 2021)). Therefore, the combined mutations do not lead to increased embryonic lethality.
Our results suggest that the double mutations in linc-9 and linc-20 do not result in a dramatic change in fertility, at least under our laboratory experimental conditions. These results still leave the open question of why these genes are expressed at high and dynamic pattern in the gonad (Ishtayeh et al., 2021; Tzur et al., 2018) if their complete removal does not lead to a change in fertility. Several answers can be suggested: It is possible that their roles are redundant with other genes with less sequence homology, or with unannotated homologous genes. Another possibility is that these transcripts’ germline expression has non-fertility related roles, or roles that manifest under different conditions as we have previously shown for linc-4 (Ishtayeh et al., 2021). Compared to protein coding genes, lincRNAs genes are very poorly evolutionary conserved (Wei et al., 2019), suggesting linc-9 and linc-20 may have been recently introduced into the genome, and not yet adopted biological roles or any biological role they used to have, has been inactivated. A recent review of the insights gained from reverse genetics experiments in lncRNAs suggested that in many cases removal of lncRNA genes lead to milder or no phenotype (Gao et al., 2020). This strengthens the possibility that indeed these genes have no functional roles. Future research may provide more insights into the functional roles of linc-9 and linc-20 expression.
Methods
Request a detailed protocolC. elegans strains
The N2 Bristol strain was utilized as the wild-type background. YBT60 was created by crossing YBT54 and YBT53 (Ishtayeh et al., 2021). All strains were cultured under standard conditions at 20 °C (Brenner, 1974). Worms were grown on NGM plates with Escherichia coli OP50 (Brenner, 1974).
Progeny quantification
The brood size was determined by placing at least 17 individual L4 worms on seeded NGM plates, transferring each worm to a new plate every 24 h, and counting their embryos and hatched progeny over 3 days. Data for Fig. 1 A, B, D, E was adapted from (Ishtayeh et al., 2021).
Statistical Analysis
Significance between mutant and wild type worms was tested by the Mann-Whitney test. Using the R package ‘wmwpow’ (Mollan et al., 2020), we calculated the power for the two-sided Wilcoxon-Mann-Whitney test performed on our two specified distributions (n=20 for WT and n=19 for linc-9: linc-20) with an empirical p-value of 0.05. We found a power of 91%.
Reagents
| Strain | Genotype | Shorten Name | Available from |
| Bristol N2 | Caenorhabditis elegans | Wild type | CGC |
| YBT53 | linc-9(huj24) | linc-9 | Tzur lab |
| YBT54 | linc-20(huj21) | linc-20 | Tzur lab |
| YBT60 | linc-9(huj24); linc-20(huj21) | linc-9; linc-20 | Tzur lab |
Acknowledgments
We thank members of the Tzur lab for their help in discussing the project. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
Funding
This work was supported by Israel Science Foundation [grant number 979/21], Ministry of Science & Technology, Israel [grant number 100594] and the Shemesh fund to Y.B.T.
Reviewed By
Jordan Ward and AnonymousHistory
Received: January 22, 2022Revision received: February 2, 2022
Accepted: February 3, 2022
Published: February 11, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Rappaport, Y; Falk, R; Achache, H; Tzur, YB (2022). linc-20 and linc-9 do not have compensatory fertility roles in C. elegans. microPublication Biology. 10.17912/micropub.biology.000524.Download: RIS BibTeX