Abstract
Fungal infection triggers the induction of antimicrobial peptide (AMP) genes in the epidermis (Pujol et al, 2008). We previously showed that this effect is suppressed by the mitochondrial unfolded protein response (UPRmt), which can be activated by knockdown of select genes including the mitochondrial metalloprotease spg-7 (Zugasti et al, 2016). Here, we confirm that RNAi against spg-7 triggers the UPRmt and blocks AMP induction during infection, whereas infection itself does not trigger the UPRmt. ATFS-1 is a key factor in the UPRmt, mediating much of the associated transcriptional response. We find that, surprisingly, ATFS-1 is not required for the suppression of AMP induction provoked by spg-7(RNAi). These data show that the mitochondrial dysfunction that blocks the immune response upon infection or wounding is independent of ATFS-1.
Description
Drechmeria coniospora is a natural fungal pathogen of Caenorhabditis elegans. Fungal spores are able to pierce the cuticle of the worm, leading to hyphal growth in the entire organism. C. elegans counters the infection by triggering a rapid innate immune response. A hallmark of the response is the secretion of antimicrobial peptides (AMP) encoded by the cnc (caenacin) family and certain nlp (neuro-peptide-like protein) genes (Dierking et al., 2011; Taffoni et al., 2020). We previously showed that impairing mitochondrial function and inducing the mitochondrial unfolded protein response (UPRmt), blocks the expression of AMP genes after infection by D. coniospora, possibly through cross-tissue signaling between the intestine and the epidermis (Zugasti et al., 2016; Ewbank & Pujol, 2016). The molecular pathways involved, however, remain unknown.
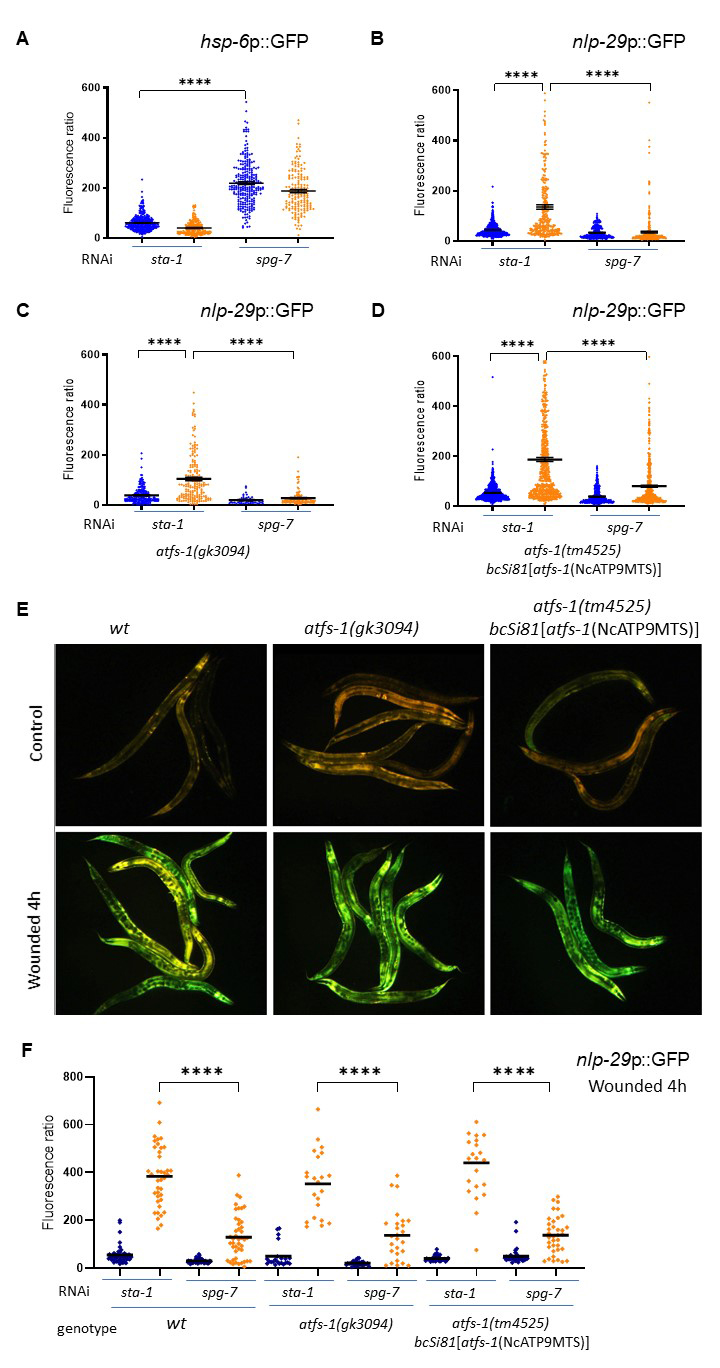
Here, we first confirmed that knock-down of the mitochondrial metalloprotease spg-7 by RNAi activates the UPRmt through the induction of hsp-6p::GFP reporter (Figure 1A) (Yoneda et al., 2004), and showed that infection by D. coniospora did not induce this reporter. Using a transcriptional reporter strain for AMP expression (nlp-29p::GFP) (Pujol et al., 2008), we also confirmed that spg-7 inactivation resulted in a robust block of AMP expression after infection compared to the sta-1 control clone (Figure 1B) (Zugasti et al., 2016).
We then investigated whether ATFS-1, a master regulator of the UPRmt, is required for the spg-7 suppression. As the responsiveness of ATFS-1 is dependent on its relatively weak mitochondrial targeting sequence (Rolland et al., 2019), its function can be impaired not only by loss-of function mutations but also by alterations that strengthen the mitochondrial targeting sequence (MTS). To examine AMP expression under both of these conditions, we crossed the nlp-29p::GFP reporter to the atfs-1(gk3094) loss of function mutant and to the atfs-1(tm4525) loss of function mutant expressing an allele with a strong MTS (bcSi81[atfs-1(NcATP9MTS)]), known to be incapable of triggering the normal UPRmt-associated transcriptional response (Rolland et al., 2019) and assayed the level of suppression of nlp-29p::GFP expression in response to spg-7(RNAi). In our hands, loss of atfs-1 did not change the induction of nlp-29 expression associated with infection by D. coniospora. Further, there was a clear suppression of AMP reporter gene expression in both mutant backgrounds upon spg-7(RNAi) following infection (Figures 1C and 1D). We also showed that atfs-1 is not required for the induction of the same immune response upon wounding (Figure 1E), nor for its suppression by spg-7 RNAi (Figure 1F). We conclude that atfs-1 is not required for the normal epidermal response to fungal infection or wounding and that the signal that provokes the repression of nlp-29 expression upon mitochondrial dysfunction is independent of the key UPRmt transcription factor ATFS-1.
Methods
Request a detailed protocolRNA interference
RNAi bacterial clones were obtained from the Ahringer library and verified by sequencing (Kamath et al., 2003). RNAi bacteria were seeded on NGM plates supplemented with 100 μg/ml ampicillin and 1 mM Isopropyl-β-D-thiogalactopyranoside (IPTG). Worms were transferred onto RNAi plates as L3 larvae to avoid the lethality associated with spg-7(RNAi) and cultured at 25 °C until young adult stage.
D. coniospora infection & wounding
Infections with D. coniospora were carried out at 25 °C as described (Pujol et al., 2008). Briefly, synchronized L4 worms obtained following treatment with an alkaline hypochlorite solution, were infected by adding 100 μl of a fresh spore solution to RNAi plates and then incubated at 25 °C for 18h. Wounding was performed with a microinjection needle in the tail region of Day1 adult worms and analysed after 4h at 25 °C as described (Taffoni et al., 2020).
Analyses with the Biosort worm sorter
Fluorescent protein expression of reporter strains was quantified with the COPAS (Complex Object Parametric Analyzer and Sorter) Biosort system (Union Biometrica; Holliston, MA) as described (Labed et al., 2008). For each strain, a minimum of 50 synchronized young adult worms were analyzed for length (assessed as TOF, time of flight), optical density (assessed as extinction) and Green and/or Red fluorescence (GFP/Red). Raw data were filtered on the TOF for adult worms (400 ≤ TOF ≤ 1000). Fluorescent ratio (Green/TOF) is presented for each worm with mean and SEM for each conditions. Statistical analyses were performed in Graphpad Prism software using one-way ANOVA with Bonferroni correction.
Reagents
IG274 frIs7[nlp-29p::GFP, col-12p::DsRed] IV (Pujol et al., 2008)
IG1825 atfs-1(gk3094) V; frIs7[nlp-29p::GFP, col-12p::DsRed] IV (This study)
IG1830 bcSi81[atfs-1(NcATP9MTS)] II; frIs7[nlp-29p::GFP, col-12p::DsRed] IV; atfs-1(tm4525) zcIs9 [hsp-60p::GFP] V (This study)
SJ4100 zcIs13[hsp-6::GFP] V (Yoneda et al., 2004)
MD4323 bcSi81[atfs-1(NcATP9MTS)] II; unc-119(ed3) III; atfs-1(tm4525) zcIs9[hsp-60p::GFP] V (Rolland et al., 2019)
VC3201 atfs-1(gk3094) V (C. elegans Deletion Mutant Consortium, 2012)
spg-7 RNAi clone sjj_Y47G6A_247.f
sta-1 RNAi clone sjj_Y51H4A.o
Acknowledgments
We thank J. Ewbank for constructive comments. Some C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Stephane Rolland for others. Worm sorting was performed by Jerome Belougne using the facilities of the French National Functional Genomics platform, supported by the GIS IBiSA and Labex INFORM.
References
Funding
This work was funded by the French National Research Agency (ANR-16-CE15-0001-01, ANR-16-CONV-0001) and institutional grants from CNRS, Aix Marseille University, National institute of Health and Medical Research (Inserm) to the CIML.
Reviewed By
AnonymousHistory
Received: January 5, 2022Revision received: January 26, 2022
Accepted: February 9, 2022
Published: February 22, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Martineau, CN; Maynard, CA; Pujol, N (2022). ATFS-1 plays no repressive role in the regulation of epidermal immune response. microPublication Biology. 10.17912/micropub.biology.000525.Download: RIS BibTeX