Graduate Program in Neuroscience, University of Illinois at Chicago
Abstract
The Caenorhabditis elegans HMX/NKX MLS-2 transcription factor was previously shown to play sequential roles in AWC general identity and the stochastic AWCON/AWCOFF subtype choice during embryogenesis. Here we analyze the expression pattern of endogenous mls-2 during AWC development using mNeonGreen (mNG) knock-in strains. Similar to transgenic GFP::MLS-2, functional mNG::MLS-2 knock-in displayed nuclear localization in AWC precursor cells but was not observed in AWC during the later embryonic stage. These results suggest that the expression of mls-2 is below the detectable level and/or the stability of MLS-2 protein is very low in AWC cells.
Description
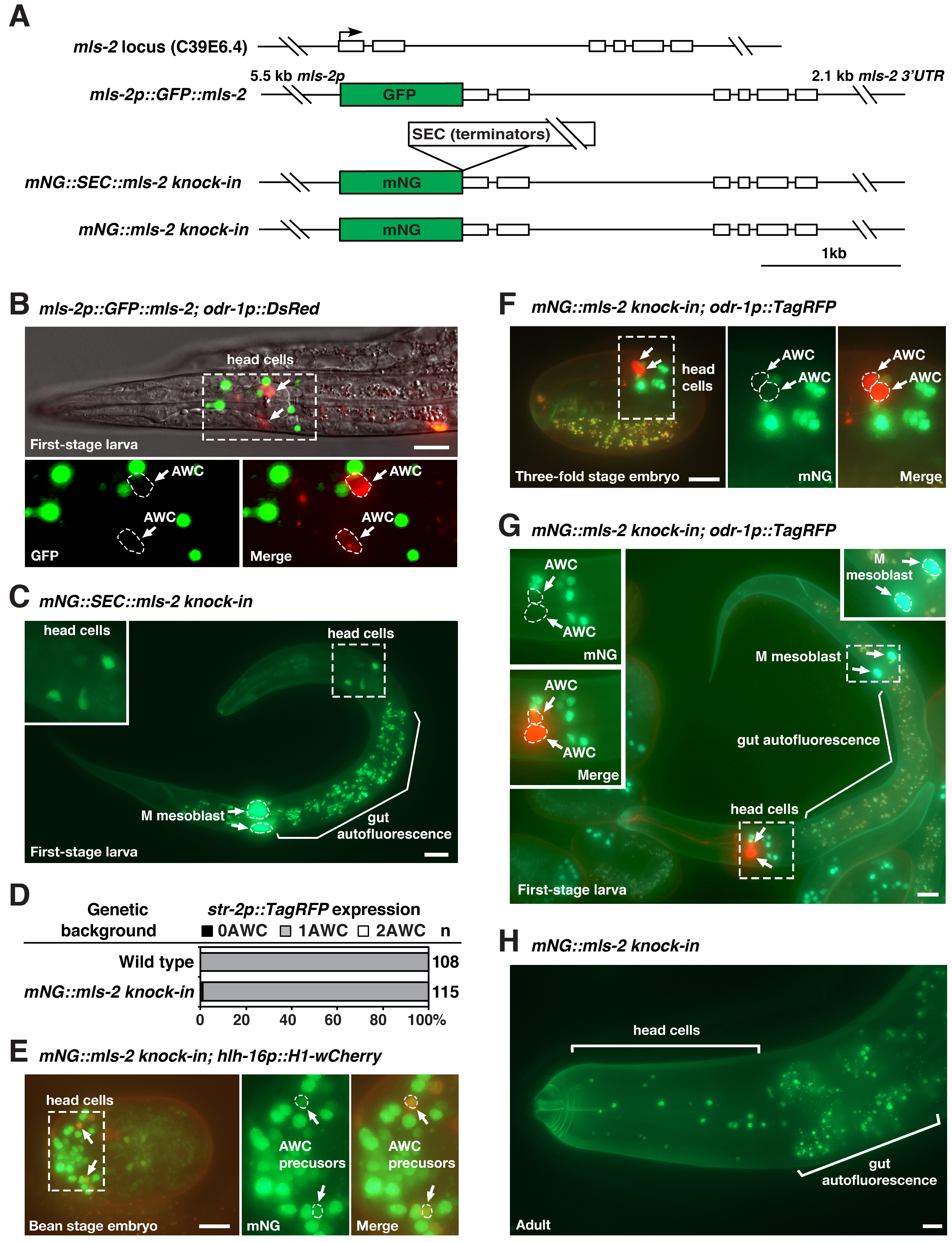
The HMX/NKX MLS-2 transcription factor plays a role in the development of the postembryonic mesoderm, CEPsh glia, tube cells of the excretory system, general AWC identity, and AWC asymmetry in C. elegans (Jiang et al., 2005; Yoshimura et al., 2008; Kim et al., 2010; Abdus-Saboor et al., 2012; Hsieh et al., 2021). The expression pattern of MLS-2 protein has been previously examined by immunohistochemical staining with anti-MLS-2 antibodies, transgenes of GFP-tagged MLS-2 (GFP::MLS-2), and mls-2::GFP fosmid reporter lines (Jiang et al., 2005; Yoshimura et al., 2008; Kim et al., 2010; Abdus-Saboor et al., 2012; Walton et al., 2015; Reilly et al., 2020) (Figure 1A). It was shown that GFP::MLS-2 was expressed in the embryonic AWC lineages from automated lineage analysis and was detected transiently in AWC neurons in first-stage larvae (Kim et al., 2010; Abdus-Saboor et al., 2012; Walton et al., 2015). However, we did not detect expression of GFP::MLS-2 transgenes in AWC neurons in late embryos or the first larval stage (Figure 1B).
To determine the expression pattern of endogenous mls-2 locus in AWC, we generated mNG::SEC::mls-2 knock-in and mNG::mls-2 knock-in animals by tagging the 5’ end of endogenous mls-2 coding region with mNG::SEC or mNG using Cas9-triggered homologous recombination (Dickinson et al., 2013; Dickinson et al., 2015; Dickinson and Goldstein, 2016) (Figure 1A). The mNG::SEC::mls-2 knock-in allele is a transcriptional reporter of mls-2, since the self-excising cassette (SEC) contains transcriptional terminators. mNG::SEC::mls-2 knock-in showed diffuse mNG expression in numerous cells in the head and the M mesoblast of first-stage larvae (Figure 1C). The mNG::mls-2 knock-in allele, generated by the SEC excision of mNG::SEC::mls-2 knock-in, is a translational reporter of MLS-2 protein. mNG::mls-2 knock-in animals displayed wild-type AWC asymmetry as determined by the expression of the AWCON marker str-2p::TagRFP (Figure 1D), suggesting that mNG::MLS-2 fusion protein is functional in AWC development. Like GFP::MLS-2 expressed from transgenes, mNG::MLS-2 knock-in was localized in the nucleus of AWC precursor cells in early embryos (Figure 1E) but was not observed in AWC cells in late embryos (Figure 1F) or early-stage larvae (Figure 1G). Our results are consistent with single-cell RNA-seq data showing that mls-2 was briefly expressed at a very low level in AWC during early embryogenesis but not detected in AWC in second-stage larvae (Cao et al., 2017; Packer et al., 2019). It was also shown that MLS-2::GFP expressed from an integrated mls-2::GFP fosmid reporter line was not detected in AWC in late larval stage or young adult-stage using NeuroPAL (Reilly et al., 2020).
Similar to MLS-2 antibody staining and GFP::MLS-2 transgenes (Jiang et al., 2005), mNG::MLS-2 knock-in was localized to the nucleus of a subset of head cells and the M mesoblast in first-stage larvae and adults (Figure 1G and 1H). mNG::SEC::mls-2 knock-in and mNG::mls-2 knock-in strains should help to determine the endogenous expression pattern of mls-2 in different cells during development.
Methods
Request a detailed protocolmNG::SEC::mls-2 and mNG::mls-2 knock-in were generated using the Cas9-triggered homologous recombination protocol as previously described (Dickinson et al., 2013; Dickinson et al., 2015).
Reagents
| Strain | Genotype | Source |
| IX1119 | oyIs44 [odr-1p::DsRed; lin-15(+)] V; vyEx535 [mls-2p::GFP::mls-2::mls-2 3’UTR (Jiang et al., 2005); ofm-1p::DsRed] | This study |
| IX4507 | mls-2(vy247 [mNG::SEC::mls-2 knock-in]) X | This study |
| IX4506 | mls-2(vy248 [mNG::mls-2 knock-in]) X | This study |
| RW10588 | unc-119(ed3); zuIs178 [his-72(1kb 5′ UTR)::his-72::SRPVAT::GFP::his-72 (1KB 3′ UTR) + 5.7 kb XbaI-HindIII unc-119(+)]; stIs10544 [hlh-16::H1-wCherry::let-858 3′ UTR] | Murray et al., 2012 |
| IX5609 | stIs10544 [hlh-16::H1-wCherry::let-858 3′ UTR] (Murray et al., 2012); mls-2(vy248 [mNG::mls-2 knock-in]) X | This study |
| IX4894 | vyIs56 [odr-1p::TagRFP] III (Cochella et al., 2014); mls-2(vy248 [mNG::mls-2 knock-in]) X | This study |
| IX3212 | vyIs68 [str-2p::TagRFP; srsx-3p::GFP] III | Cochella et al., 2014 |
Acknowledgments
We thank Dr. Jun Liu for the plasmid mls-2p::GFP::mls-2::mls-2 3’UTR. The RW10588 strain was provided by the Caenorhadbitis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
Funding
This work was funded by a grant, R01GM098026 awarded to CFC, from the National Institute of General Medical Sciences of the National Institutes of Health.
Reviewed By
Paschalis Kratsios and Jayson SmithHistory
Received: January 14, 2022Revision received: February 9, 2022
Accepted: February 10, 2022
Published: February 22, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Xiong, R; Hsieh, YW; Chuang, CF (2022). mNG-tagged mls-2 knock-in alleles in C. elegans. microPublication Biology. 10.17912/micropub.biology.000529.Download: RIS BibTeX