Department of Molecular Biosciences, Northwestern University, Evanston, IL, 60208, USA
Description
Lee et al., 2017 showed that CB4856 has a lower nictation ratio (the number of nictating dauers over the number of moving dauers on a micro-dirt chip) than N2, and that a genetic locus (nict-1) on chromosome IV mediates this phenotype difference. Although this paper had no evidence of any other genetic loci for nictation behavior, we have studied other quantitative trait loci (QTL) using different linkage-based mapping strategies. Here, we report another genetic locus that was identified using introgression lines on chromosome V.
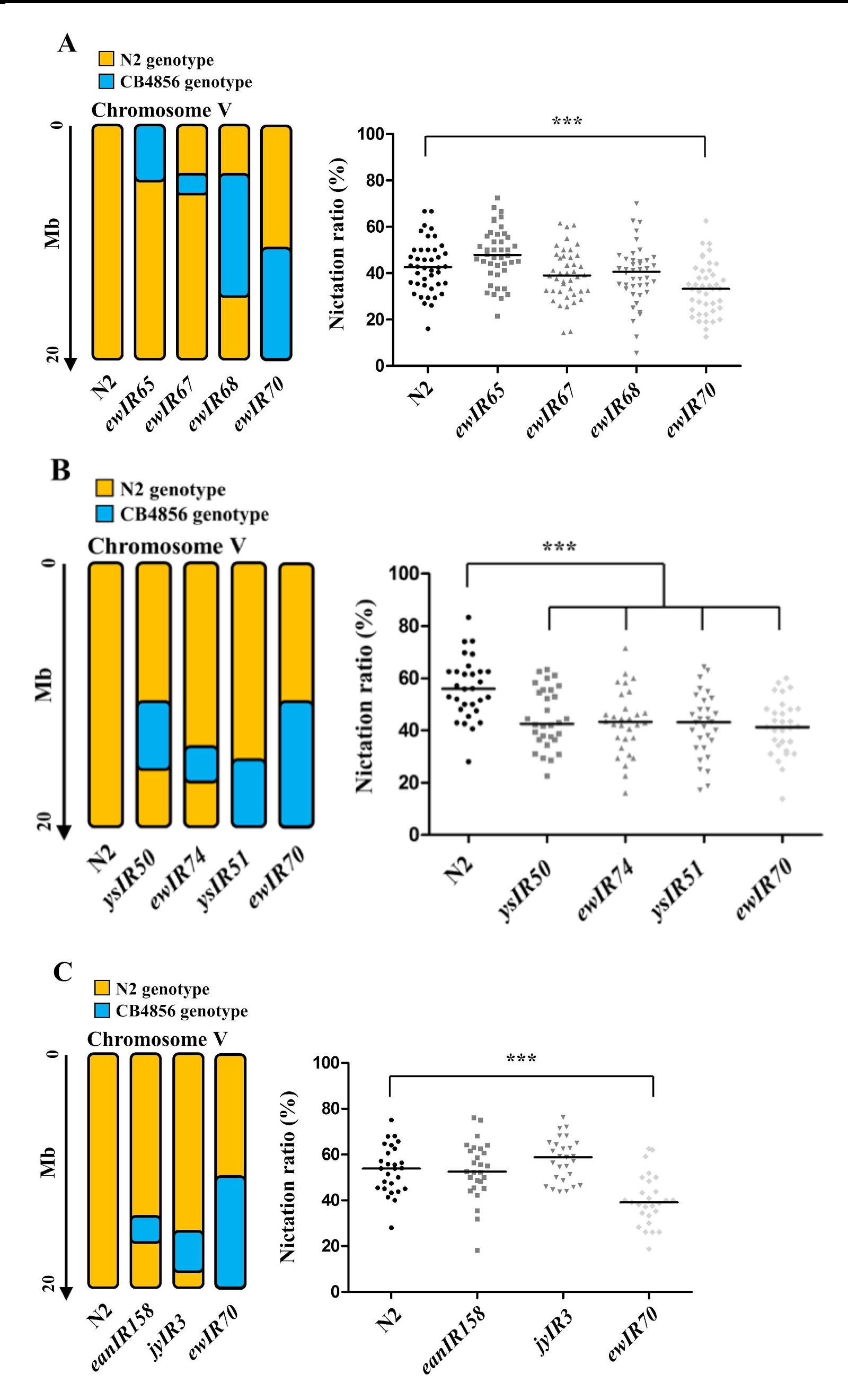
N2 x CB4856 introgression lines were tested using a population nictation assay (Lee et al., 2015). We measured the nictation ratios of the ewIR65, ewIR67, ewIR68, and ewIR70 lines, which contain N2-type genome in all chromosomes except for CB4856-type chromosome V segments (Figure 1A). Among them, only ewIR70 line showed significant lower nictation ratio. Therefore, we narrowed down the chromosome V right region.
We also measured nictation ratios of ysIR50, ewIR74, and ysIR51 lines (Figure 1B). ysIR50 and ysIR51 were derived from N2 x ewIR70. ysIR50, ewIR74, and ysIR51 lines showed significantly lower nictation ratios than that of N2. Because the putatively responsible QTL region is covered by another near-isogenic region, eanIR158, which was made independently by Erik Andersen’s laboratory, we measured the nictation ratios of eanIR158 and jyIR3 lines and found that they did not show any significant difference from N2 (Figure 1C).
eanIR158 and jyIR3 lines were built with QX1430 x N2 (personal communication) and QX217 x N2 (Balla et al., 2015), respectively, and QX1430 and QX217 were made by Leonid Kruglyak’s laboratory (Andersen et al., 2015; Rockmen and Kruglyak, 2009). However, ewIR70 and ewIR74 were built by Jan Kammenga’s laboratory (Doroszuk et al., 2009). Thus, we concluded that the two laboratories may have used different N2 or CB4856strain derivatives, which may have altered genetic variations in the introgression lines. A take-home lesson is that it is important to examine NILs derived from common original strains in order to follow QTL without being affected by differences in background genotypes.
Jan Kammenga kindly provided their N2 x CB4856 introgression lines for chromosome V, which include ewIR65, ewIR67, ewIR68, ewIR70, and ewIR74, and Erik Andersen also gave us many introgression lines, which include eanIR158 and jyIR3 covering 15-19 Mb of chromosome V. We appreciate their help.
Reagents
Strains: N2, LJ1250 ysIR50 (V, CB4856>N2), LJ1251 ysIR51 (V, CB4856>N2), ECA238 eanIR158 (V, CB4856, N2), and ERT248 jyIR3 (V, CB4856>N2).
Genotypes: ewIR65 (V, CB4856>N2), ewIR67 (V, CB4856>N2), ewIR68 (V, CB4856>N2), ewIR70 (V, CB4856>N2), and ewIR74 (V, CB4856>N2).
References
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (No. 2016K1A3A1A47921615). Jun Kim was supported by POSCO Science Fellowship from the POSCO TJ Park Foundation.
Reviewed By
Erik AndersenHistory
Received: August 24, 2017Accepted: September 13, 2017
Published: September 19, 2017
Copyright
© 2017 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Kim, J; Lee, D; Lee, J (2017). A quantitative trait locus for nictation behavior on chromosome V. microPublication Biology. 10.17912/W23D39.Download: RIS BibTeX