Description
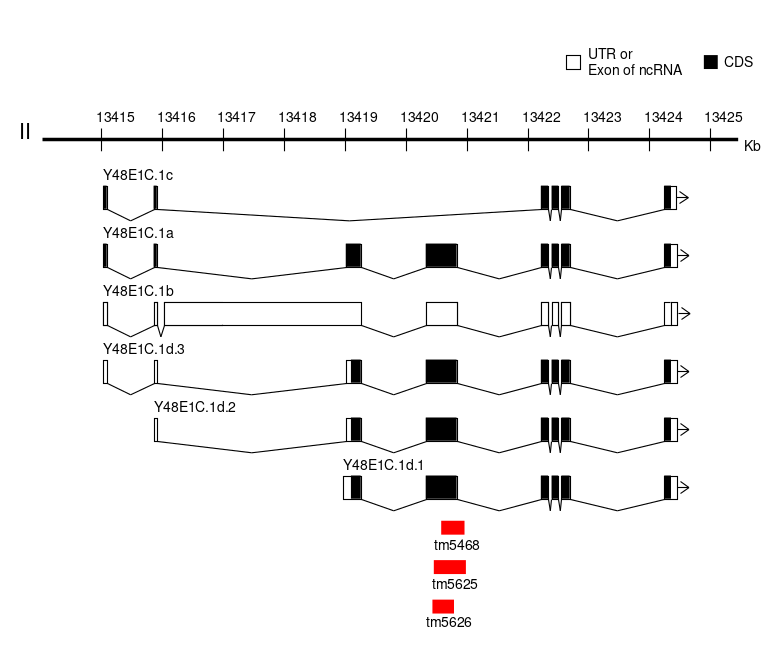
We report tm5468, tm5625 and tm5626 as novel deletion alleles of the gene Y48E1C.1 that is the only ortholog of human calmodulin-lysine N-methyltransferase (CAMKMT)1. CAMKMT encodes an evolutionarily conserved enzyme class I protein methyltransferase that acts in the formation of trimethyllysine in calmodulin for calcium-dependent signaling2. CAMKMT mutation is associated with Hypotonia-cystinuria syndrome in human2,3. The alleles were isolated from the comprehensive screening of gene deletions generated by TMP/UV4. In the screening, all the alleles were detected by nested PCR using the following primer sets, 5’- TCAAGCCACGCCCACACTTA-3’ and 5’- GAAGGCATACAGTGGGGGTA-3’ for the first round PCR and 5’- CGCCCACACTTAATGGTTAT-3’ and 5’- GGGCAGTGTAGGGATACTGT-3’ for the second round PCR. By Sanger sequencing, the 30 bp flanking sequences of the alleles tm5468, tm5625 and tm5626 were identified as AATCCTTCACACACCACAACAGAAATCCTA – [384 bp deletion] -CGAGGTCACGCCCACACATTGGGCGGAGTT, CCGATGCTCCGTGCTGCTCCAAGTGCTCCG – [627 bp deletion + 9 bp insertion (TAATCTTGT)] – AGTACTCCTACAGTATCCCTACACTGCCCC, and AAAAAAGGATGACGTCACAGTTGCTCCGAT – [256 bp deletion] – ACGCCGATTCGGCAGCCGAATGATCTACAG, respectively. Based on the information about the splicing isoforms of Y48E1C.1 (WormBase, http://www.wormbase.org, WS259), the forth exon of Y48E1C.1a, Y48E1C.1b (annotated as non cording RNA) and the second exon of Y48E1C.1d transcripts are deleted in tm5468, tm5625 and tm5626(Fig. 1). Presumably, all of the alleles do not affect Y48E1C.1c. According to information of protein in Wormbase, this exon contains a predicted some motif, suggesting hypothetical functional deficiency of Y48E1C.1aY48E1C.1b, and Y48E1C.1d in the deletion mutants. In addition, these alleles are expected to be usable for comparing functions among the isoform c and the other isoforms. However, no visually obvious phenotypes (Let, Unc, and Dpy) were observed in tm5468, tm5625 and tm5626.
References
Funding
The National BioResource Project.
Reviewed By
James LeeHistory
Received: August 30, 2017Accepted: October 3, 2017
Published: October 3, 2017
Copyright
© 2017 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Hori, S; Suehiro, Y; Yoshina, S; Mitani, S (2017). Novel deletion alleles of a C. elegans gene Y48E1C.1, named as tm5468, tm5625 and tm5626. microPublication Biology. 10.17912/W2CQ14.Download: RIS BibTeX