Interdisciplinary Biological Sciences Program, Northwestern University, Evanston, IL, 60208, USA
Abstract
To better understand the mechanism of resistance caused by putative interactions between beta-tubulin and benzimidazole compounds, we sought to purify nematode-specific beta-tubulins using heterologous expression after replacement of the single Saccharomyces cerevisiae beta-tubulin gene. However, we found that haploid yeast cells containing nematode-specific beta-tubulin genes were not viable, suggesting that nematode beta-tubulin cannot substitute for the loss of the yeast beta-tubulin gene.
Description
Parasitic nematode infections continue to have an enormous impact on human and livestock health worldwide (Hotez et al., 2014; Kaplan & Vidyashankar, 2012). A limited arsenal of anthelmintic drugs are available to combat these infections. One of the most widely used classes is benzimidazoles (BZ), and resistance against this class is widespread (Kaplan & Vidyashankar, 2012). Previous studies to understand parasitic nematode resistance using the free-living model organism Caenorhabditis elegans showed that variation in the C. elegans beta-tubulin gene ben-1, an ortholog of beta-tubulins in parasitic nematodes, confers resistance to BZ drugs (Dilks et al., 2020; Driscoll et al., 1989; Hahnel et al., 2018). The most common missense mutation resistance alleles are F167Y, E198A, and F200Y (Avramenko et al., 2019; Mohammedsalih et al., 2020). Although computational models have predicted that these amino acids are involved in the binding of BZ compounds to beta-tubulins, the binding remains to be investigated empirically at the structural level because nematode-specific beta-tubulin structures have not been created (Aguayo-Ortiz et al., 2013; Hahnel et al., 2018). To better understand the mechanisms of resistance, we sought to obtain those crystallographic structures.
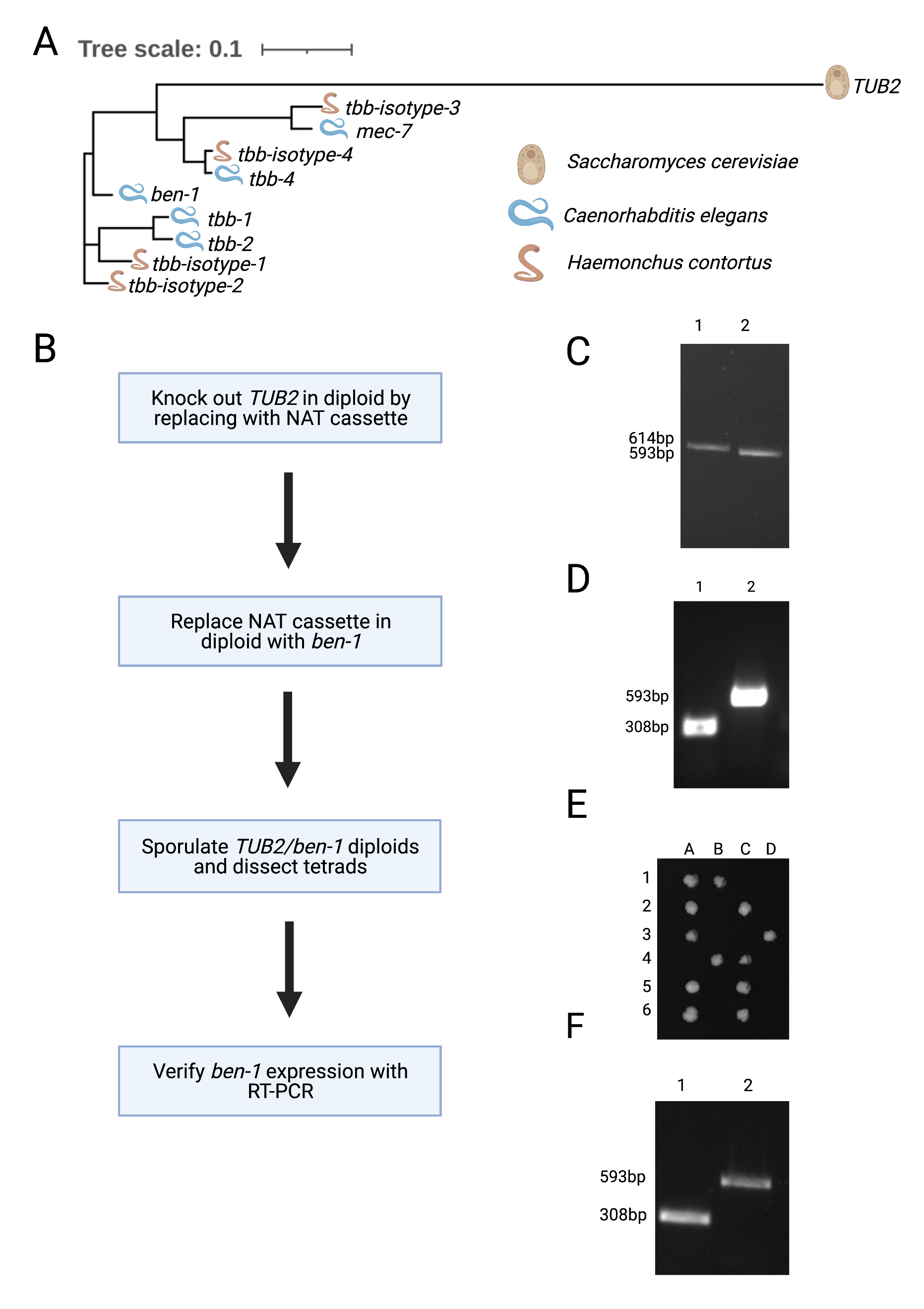
To isolate BEN-1 for crystallographic studies, tags would permit the purification of this beta-tubulin from the other five beta-tubulins in C. elegans. Unfortunately, endogenous tags of BEN-1 eliminate its function (Dilks and Andersen, unpublished results). Because of this limitation, we turned to expression of three different versions of nematode beta-tubulins, C. elegans BEN-1 and two beta-tubulin isotypes from the parasitic nematode Haemonchus contortus, in Saccharomyces cerevisiae, which has a single beta-tubulin gene (Figure 1A).
We used the following procedure to replace the S. cerevisiae beta-tubulin gene with each of the nematode beta-tubulin genes (Figure 1B). These replaced beta-tubulin genes were expressed using the native S. cerevisiae promoter. First, TUB2 was deleted in diploid BY4743 cells and replaced with the Nourseothricin N-acetyl transferase (NAT) cassette selection marker (Hansen et al., 2003). Successful deletion of one copy of the TUB2 coding sequence was verified using colony PCR of a sample that grew in the presence of NAT (Figure 1C). The NAT cassette was then substituted with yeast codon-optimized C. elegans ben-1 along with the HIS3 selection marker (Burke & Gould, 1994). Successful replacement of ben-1 at the TUB2 locus was verified using colony PCR on samples that grew in histidine-deficient conditions (Figure 1D). We sporulated diploids of the ben-1 replacement strain then dissected tetrads to identify haploid cells that contained the C. elegans beta-tubulin gene ben-1. We found that dissected tetrads gave rise to only two viable haploid cells instead of four when grown on complete media (YPD) (Figure 1E). The same result was obtained after replacement of TUB2 with either of the two H. contortus beta-tubulin isotype genes (data not shown). The two surviving colonies were also unable to grow in histidine-deficient conditions, suggesting that replacement of TUB2 with nematode beta-tubulin genes is lethal. Using RT-PCR of the heterozygous diploid strain with one copy of TUB2 and one copy of ben-1 at the TUB2 locus, we found that ben-1 mRNA was expressed (Figure 1F), suggesting that the nematode beta-tubulin is deficient at protein expression, lacks proper stability, and/or cannot function with the yeast alpha-tubulin gene. Our results indicate that nematode beta-tubulin genes can not substitute for the yeast beta-tubulin gene TUB2.
Beyond the phylogenetic difference between nematode and yeast beta-tubulins, it has been observed that the dynamics of microtubules in C. elegans greatly differ from other eukaryotes (Chaaban et al., 2018), suggesting that nematode beta-tubulins might be unsuitable replacements for yeast beta-tubulins. It is possible that both alpha- and beta-tubulin might need to be replaced by their nematode-specific versions to obtain viable yeast and allow for the characterization of benzimidazole and beta-tubulin binding to better understand the mechanism of resistance.
Methods
Request a detailed protocolStrain Construction
S288C derivative strains BY4741 and BY4742 (Brachmann et al., 1998) were mated to make diploid BY4743.
Plasmid Construction
pFA6-NAT and pKT128
The construction for the plasmids containing the NAT cassette, pFA6-NAT, and the S. pombe HIS3 marker, pKT128, have been outlined previously (Janke et al., 2004; Sheff & Thorn, 2004).
pECA101, pECA103, and pECA103
Yeast codon optimized sequence for C. elegans ben-1 and H. contortus tbb-isotype-1 were cloned into a pUC57 vector and H. contortus tbb-isotype-2 was cloned into a pJET1.2 vector by GenScript (Piscataway, NJ) to make pECA101, pECA102, and pECA103 respectively. The plasmid was cloned into DH-alpha competent cells and prepped with a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany).
Yeast Transformation
Cells were grown overnight in YPD at 24°C, diluted to an OD of 0.15 and then grown to an OD of 0.5-1.0. Cells were resuspended in 100 mM Lithium acetate and combined with 50% PEG, 1M Lithium acetate, 10 mg/mL salmon sperm DNA, water, and the desired amplicon for integration. After incubating at 30°C for 30 minutes, DMSO was added and then reactions were heat shocked at 42°C for 15 minutes. Cells were washed and either recovered overnight in YPD and plated on YPD with NAT for NAT selection or resuspended in water and plated immediately on -HIS plates for HIS3 selection. Plates were incubated for 48 hours at 30°C.
TUB2 Knock-out
A NAT cassette sequence flanked by the beginning and end of the S. cerevisiae TUB2 sequence was amplified with pFA6-NAT and the following oligonucleotides (IDT, Coralville, IA)
oECA1756: ATGAGAGAAATCATTCATATCTCGACAGGTCAGTGTGGTACGGATCCCCGGGTTAATTAA
oECA1757: TTATTCAAAATTCTCAGTGATTGGTTCATCTTGGTTTTGTGAATTCGAGCTCGTTTAAAC
Oligonucleotides to verify NAT cassette replacement (IDT, Coralville, IA)
oECA1758: AACTGGTGCACTTAATCGCTG
oECA1759: CAATTCAACGCGTCTGTGAGG
Oligonucleotides for mating type positive control (IDT, Coralville, IA)
MAT-F: TTACTCACAGTTTGGCTCCGGTGT
MAT-R: GAACCGCATGGGCAGTTTACCTTT
Ben-1 replacement
A codon-optimized C. elegans ben-1 sequence flanked by sequence upstream and downstream of TUB2 was amplified with pECA101 and the following oligonucleotides (IDT, Coralville, IA)
oECA1762: CTACTACAACTACAAAAGCAAAATCTCCACAAAGTAATATAATGAGAGAAATTGTTCATG
oECA1763: CATAAGAAATTCGCTTATTTAGAAGTGGCGCGCCTTATTCAGCGTCACCATC
An S. pombe HIS3 selective marker was amplified with pKT128 and the following oligonucleotides (IDT, Coralville, IA)
oECA1764: GATGGTGACGCTGAATAAGGCGCGCCACTTCTAAATAAGCGAATTTCTTATG
oECA1765: AGAGAAGAAGAAAGGTAAGAAAAAGAAAGGAAAGCAACTTAATCGATGAATTCGAGCTCG
Oligonucleotides to confirm the substitution or expression of ben-1 (IDT, Coralville, IA)
oECA1769: CATACAATGCTACATTGTCAG
oECA1770: CAAAGCTCTATATGCTTGAG
Sporulation and Tetrad dissection
Cells were grown overnight in YPD at 24°C and then nitrogen starved in SPO Media at 24°C for 5-6 days until sporulated. Cells were then digested in 0.5 mg/mL zymolyase, resuspended in 1.2M sorbitol, and tetrads were dissected onto YPD plates and incubated for 48 hours at 30°C. Grown colonies were then struck on to -HIS plates and incubated overnight at 30°C.
RNA extraction and RT-PCR
Cells with ben-1 substitution were grown to an OD600 of 10 overnight in YPD at 24°C and then pelleted and resuspended in High Salt RNA Buffer (0.3 M NaCl 20 mM Tris pH 8 10 mM EDTA 1% SDS), then separated with TE-saturated Phenol:Chloroform:Isoamyl alcohol. RNA was then extracted with Chloroform, precipitated with ethanol, and resuspended in water.
RNA was converted to cDNA using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). The ben-1 verification and mating-type sequences were then amplified.
Acknowledgments
We would like to thank Conor Lee-Smith for his contributions in strain construction. We would also like to thank Janneke Wit and Clayton Dilks for their helpful suggestions for the manuscript.
References
Funding
This work was supported by the National Institutes of Health NIAID R01 AI153088
Reviewed By
James WasmuthHistory
Received: June 7, 2021Revision received: June 19, 2021
Accepted: June 21, 2021
Published: June 30, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Gibson, SB; Harper, CS; Lackner, LL; Andersen, EC (2021). The Caenorhabditis elegans and Haemonchus contortus beta-tubulin genes cannot substitute for loss of the Saccharomyces cerevisiae beta-tubulin gene. microPublication Biology. 10.17912/micropub.biology.000411.Download: RIS BibTeX