School of Biological Sciences, University of Auckland, Auckland, New Zealand
Description
The entire C. elegans intestine is derived from a single endodermal progenitor cell (E), the posterior daughter arising from the asymmetric division of the EMS blastomere. During early embryonic development, maternally provided SKN-1/Nrf2 activates the mesendoderm gene regulatory network (GRN) in both E and its sister, MS. A triply redundant Wnt/MAPK/Src signaling system from the neighboring P2 blastomere polarizes EMS, resulting in activation of E fate on the side contacting it. In MS, and in an unsignaled E cell, POP-1/Tcf represses expression of the redundant endoderm specifying factors, the END-1 and -3 GATA-type transcription factors. In a normal E cell, Wnt (initiated by the MOM-2/Wnt ligand) and MAPK signaling (through the MOM-4 MAPKKK) converge on POP-1 to convert it from a repressor to an activator of the end genes which, in collaboration with SKN-1, activates E cell fate (Thorpe et al. 1997; Maduro and Rothman 2002; McGhee 2007; Maduro 2017).
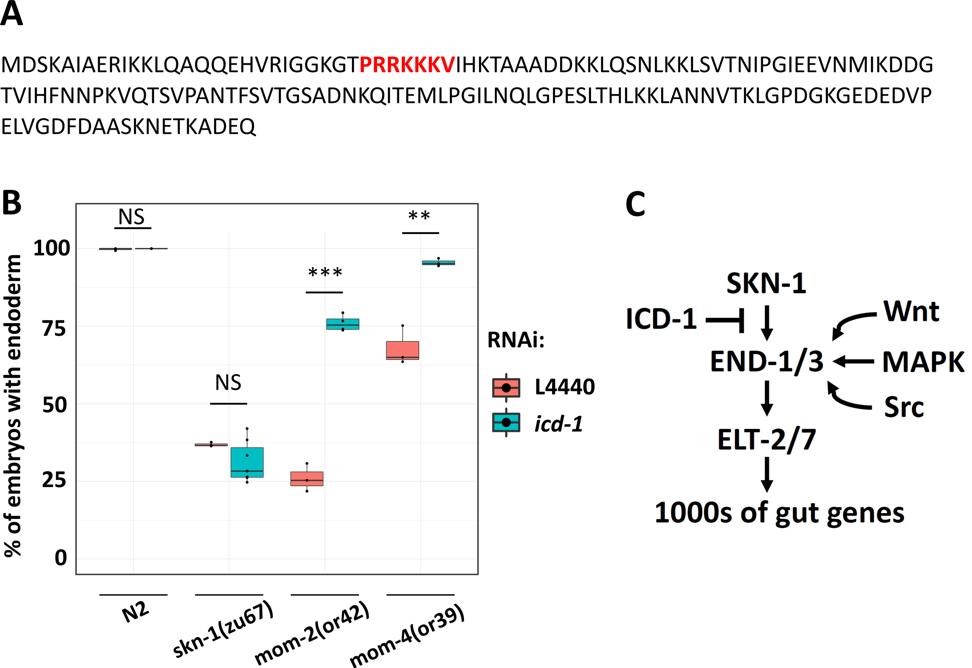
Basal transcription factor 3 (BTF3) facilitates transfer of nascent polypeptide chains into mitochondria and regulates transcription in plants and animals (Jamil et al. 2015). We previously found that the C. elegans BTF-3 orthologue, ICD-1, is required to prevent apoptosis: eliminating icd-1 leads to increased cell death in embryos and larvae (Bloss et al. 2003). However, consistent with its potential function as a transcription factor, ICD-1 contains a putative nuclear localization signal in the N-terminus (Fig.1A) (Lange et al. 2007). Here, we report that ICD-1 performs a function in endoderm specification. We found that icd-1 RNAi does not affect endoderm specification in a wild-type N2 background or the gut-less phenotype of skn-1(-) embryos. However, knockdown of icd-1 strongly suppresses the absence of gut in mom-2/Wnt(-) embryos (mom-2(or42): 26.0% ± s.d. 4.5% with gut vs. mom-2(or42); icd-1(RNAi): 75.9% ± s.d. 2.6%). Similarly, depleting ICD-1 rescues the gut-less phenotype of mom-4/Tak1(-) embryos (mom-4(or39): 67.9% ± s.d. 6.3% with gut vs. mom-4(or39); icd-1(RNAi): 95.5% ± s.d. 1.2%) (Fig. 1B). Our findings suggest a model in which ICD-1 antagonizes the SKN-1 input, perhaps by preventing SKN-1 from binding to end-1/3 promotors. This possibility is also consistent with the finding that competition between ICD-1 and SKN-1 is seen in the context of the unfolded protein response (UPR). SKN-1 binds to and activates hsp-4, which codes for an endoplasmic reticulum chaperone BiP (Glover-Cutter et al. 2013). Depleting ICD-1 results in upregulation of hsp-4 and activation of the UPR (Arsenovic et al. 2012), suggesting that ICD-1 and SKN-1 perform opposing functions in other contexts. In this hypothesized model, ICD-1 may act to fine-tune developmental signals, thereby ensuring proper specification and differentiation of endoderm (Fig. 1C).
Reagents
Strains
JJ185 dpy-13(e184) skn-1(zu67) IV; mDp1 (IV;f)
JR3936 dpy-13(e184) skn-1(zu67) IV/nT1 [qIs51] (IV;V)
EU384 dpy-11(e1180) mom-2(or42) V/nT1 [let-?(m435)] (IV;V).
EU414 unc-13(e1091) mom-4(or39)/hT2 I; +/hT2 [bli-4(e937) let-?(h661)] III.
RNAi and quantification of endoderm specification
E. coli HT115 expressing icd-1 dsRNA was obtained from the Ahringer RNAi library (Kamath et al. 2003). RNAi experiments for embryonic endoderm specification were performed as described (Torres Cleuren et al. 2019). In brief, bacteria were grown at 37oC in LB containing 50 μg/ml ampicillin. The overnight culture was then diluted 1:10. After 4 hours of incubation at 37oC, 1 mM of IPTG was added and 60 μl was seeded onto 35 mm agar plates containing 1 mM IPTG and 25 μg/ml carbenicillin. Seeded plates were allowed to dry overnight before use. 20-30 L4 or young adults were placed on the seeded RNAi plate. 24 hours later, they were transferred to a fresh RNAi plate and allowed to lay eggs for four hours. The adults were then removed, leaving the embryos to develop for an extra 5-7 hours. Embryos expressing birefringent gut granules were quantified and imaged on an agar pad using a Nikon Ti-E inverted microscope under dark field with polarized light (Clokey and Jacobson 1986; Hermann et al. 2005). All experiments were performed at 20oC.
Acknowledgments
Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The authors thank members of Rothman lab, especially Pradeep M. Joshi, for helpful advice and feedback.
References
Funding
This research was supported by NIH grants # HD081266 and HD082347 to J.H.R.
Reviewed By
AnonymousHistory
Received: September 3, 2019Accepted: September 26, 2019
Published: October 4, 2019
Copyright
© 2019 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ewe, CK; Torres Cleuren, YN; Alok, G; Rothman, JH (2019). ICD-1/BTF3 antagonizes SKN-1-mediated endoderm specification in Caenorhabditis elegans. microPublication Biology. 10.17912/micropub.biology.000167.Download: RIS BibTeX