Department of Biochemistry, Purdue University, West Lafayette, IN
Department of Pathology, University of Texas Medical Branch, Galveston, TX
Description
T-DNA tagging is a method to generate mutations in plants by random insertion. It is an important tool for the study of gene function in Arabidopsis because it allows you to see how the plant responds when expression of a specific protein is altered. T-DNAs carrying promoter elements that can cause transcriptional activation, called activation tags, have previously been shown to be effective at identifying novel genes (Tani et al. 2004). We attempted to develop a gene silencing T-DNA, by modifying the existing microRNA-induced gene silencing (MIGS) platform (Han et al. 2015). In this platform, the transgene consists of a promoter driving expression of a miR173 target sequence directly adjacent to the gene sequence to be silenced (Zhang 2014). Expression produces an mRNA which is bound by the complementary, naturally occurring Arabidopsis microRNA, miR173. This induces the production of small interfering RNAs (siRNAs) which target homologous transcripts for degradation (Zhang 2014).
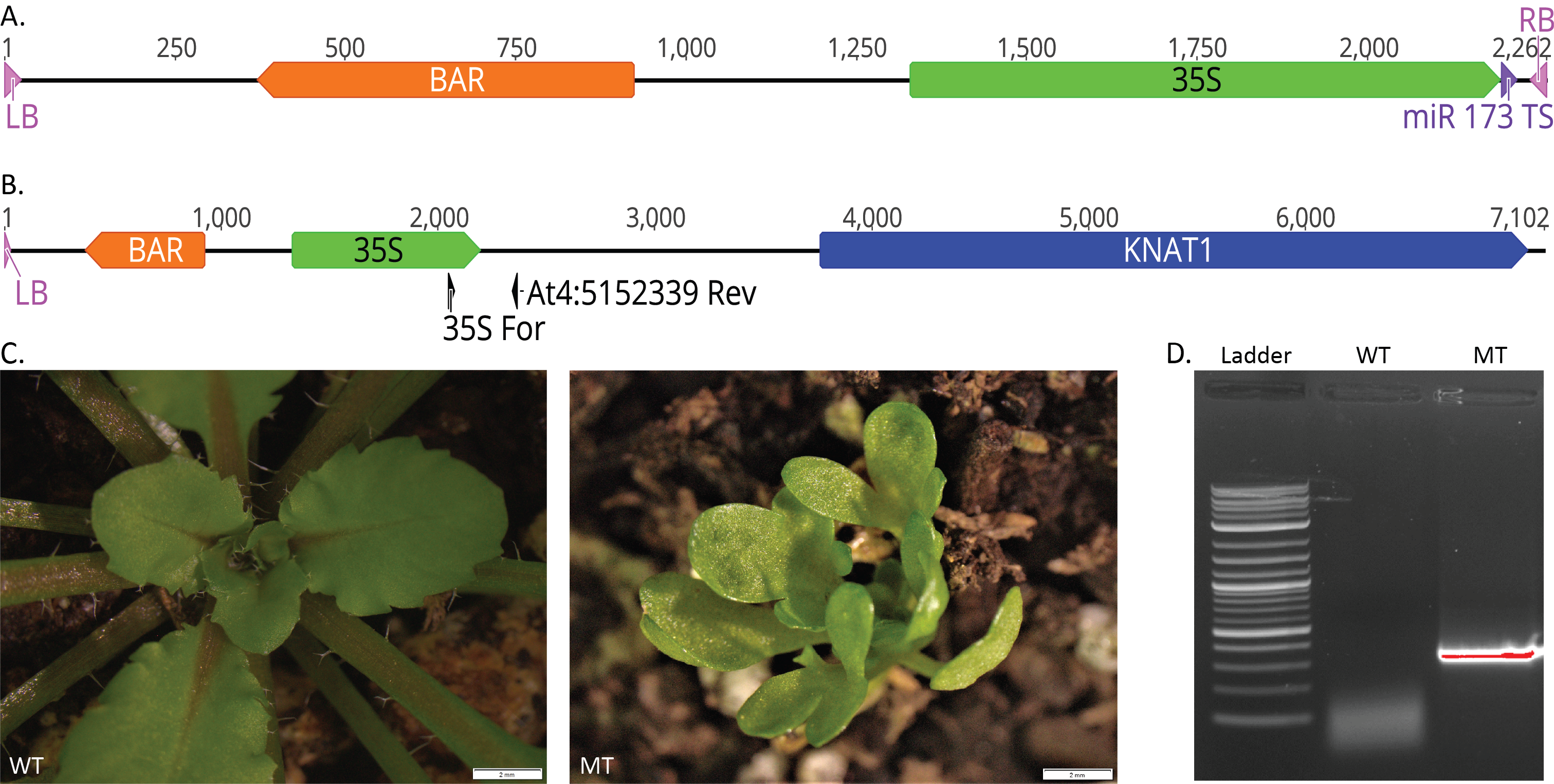
The T-DNA transgene containing the BAR gene, a CaMV 35S promoter, and the miR173 target sequence was made by modifying the existing pMIGS vector (Han et al. 2015) (Fig. 1A). The T-DNA transgene was transformed into wild-type Arabidopsis thaliana plants using the floral dip method (Clough et al. 1998). Screening for mutant phenotypes in the offspring, we identified one plant, named Gamora, which exhibited altered leaf shape, delayed flowering, and a reduced seed set in a dominant manner (Fig. 1C). Whole genome sequencing of the Gamora mutant using Oxford Nanopore MinION sequencing (Michael et al. 2018) produced 111,593 reads with an average length of 6 kb, resulting in approximately 670 Mb of sequence. One read contained the T-DNA sequence along with adjacent Arabidopsis genome sequence, allowing us to determine where the transgene had inserted in the genome (Fig. 1B). The T-DNA is inserted upstream of KNAT1 (At4G08150), likely in the promoter region. PCR amplification (Fig. 1D) and sequencing of the transgene/genome junction site revealed that the T-DNA right border and part of the miR173 target sequence were missing, but the CaMV 35S promoter was still intact (Fig. 1B). Lacking these sequences, the T-DNA is likely to function as an activation tag because the CaMV 35S promoter has been shown to induce the expression of nearby genes (Odell et al. 1985). KNAT1 has been identified previously and is known to encode for a KN1-like homeodomain protein which is primarily localized in the shoot apical meristem in Arabidopsis (Lincoln et al. 1994). Overexpression of the maize homolog for KNAT1 (Kn1) under control of the 35S promoter in tomato resulted in dwarfed and bushy plants (Hareven et al. 1996). It has also been shown that overexpression of Kn1 and KNAT1 under control of the 35S promoter in Arabidopsis results in plants with highly lobed leaves, defects in floral development, reduced fertility, slow growth, and dominant inheritance, similar to the Gamora phenotype (Lincoln et al. 1994). Together, this suggests that the Gamora phenotype is due to overexpression of the KNAT1 gene.
The results obtained from this project provide insight into the development and use of Arabidopsis gene discovery tools. Though this study identified a mutant phenotype and its underlying gene, it also suggests that there are limitations to our MIGS-based silencing tagging design. Our strategy requires that the miR173 target sequence be placed near the ends of the T-DNA, which increases the probability for loss of this sequence upon insertion. Afolabi et al. (2004) found that non-intact T-DNAs were present in over 70% of transgenic rice lines, in most cases reflecting loss of the mid to right border portion of the T-DNA. This suggests that we may have better success with integration of the miR173 target sequence if we include more sequence between the target sequence and the right border. Our study also indicates that Nanopore sequencing can be successfully utilized to identify transgene locations in Arabidopsis.
Methods
Request a detailed protocolMIGS Transgene Design and Plant Transformation
The pEG100 MIGS T plasmid was produced by transferring the EcoRI to XbaI fragment from pMIGS (Han et al. 2015) to pEarleyGate100 (Earley et al. 2006). The resulting plasmid was then digested with PmeI and Xbal, blunted with T4 polymerase, and ligated to delete the region between the target site and the right border. Wild-type Arabidopsis thaliana plants were transformed using the floral dip method as described by Clough et al., 1998. The transgenic offspring were selected using Basta herbicide.
Oxford Nanopore MinION sequencing
DNA was extracted from 15 pooled mutant plants using the CTAB method (Liu et al. 1995) and then purified using the E.Z.N.A Plant DNA purification kit (Omega Bio-tek, Norcross, GA). The DNA library was prepared with the ONT Ligation Sequencing Kit 1D (Oxford Nanopore Technologies, Oxford Science Park, UK) by the Functional Genomics Core at the University of South Carolina according to the recommended protocol (Michael et al. 2018). The library was sequenced using the Nanopore R9.4 Spot-On Flow cell for 24 hours. Geneious Software was used to BLAST the reads against the transgene sequence.
PCR
PCR analysis was performed on wild-type Arabidopsis (WT) and Gamora mutant (MT) DNA with 2X Taq RED Maser Mix, 1.5 mM MgCl2, and 35S For (AGACGTTCCAACCACGTCTTCAAAGCAAG) and At4:5152339 Rev (TGCATTCGAAATGTTTTCTTTTCC) primers flanking the region between the CaMV 35S promoter and AtG08151 (Fig. 1B). The PCR reaction was a 10 μl reaction that included a 4 min denaturation at 95°C, then 30 cycles of (30 sec., denaturation at 95°C, 30 sec annealing at 58°C, and a 1 min 30 sec extension at 72°C) and a final extension time of 7 min at 72°C.
Acknowledgments
We would like to thank Jacob Reagin for technical assistance with experiments and Priscilla Redd for help with editing.
References
Funding
This project was funded by the University of South Carolina Magellan Scholar Program and the National Science Foundation Award #1444581.
Reviewed By
Beth KrizekHistory
Received: April 1, 2020Revision received: May 18, 2020
Accepted: May 19, 2020
Published: May 21, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Moulton, K; Diaz, S; Strother, A; Hancock, CN (2020). A partial T-DNA insertion near KNAT1 results in lobed Arabidopsis thaliana leaves. microPublication Biology. 10.17912/micropub.biology.000253.Download: RIS BibTeX