Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, Rua Campo Alegre, 4169-007 Porto, Portugal
Description
Hexokinases (HXKs) are central enzymes in the carbohydrate metabolism, by controlling the entry point of glucose into glycolysis, while also displaying regulatory roles. In plants, Arabidopsis thaliana HXK1 (AtHXK1, also known as AtGIN2) is involved in hormonal signalling, which likely explains its involvement in plant growth and development (Moore et al., 2003). One interesting feature of AtHXK1 is that, in the presence of exogenous glucose, it localizes in the nucleus to form a complex that inhibits photosynthesis-associated transcripts (Cho et al., 2006). This feature is central to cope with excess sugar by diminishing sugar biosynthesis. With such a central role, it is likely that AtHXK1 has several layers of regulation, and it is therefore logical that AtHXK1 may be targeted by different posttranslational modifications (PTMs). In support, PTMs such as phosphorylation and ubiquitination have been proposed to control HXK activity in human cells (Zhang et al., 2017; Lee et al., 2019). At the plant level however, the knowledge concerning HXK regulation is very scarce.
In a previous work, it was reported that Arabidopsis HXK1 interacts with Arabidopsis SUMO E2 conjugase (SCE1), based on a SCE1-bait yeast-two-hybrid screening assay (Elrouby and Coupland, 2010). This places HXK1 as a potential target for the peptide small ubiquitin-like modifier (SUMO). Conversely, we genetically demonstrated, using null mutants for HXK1 and the central SUMO pathway component SIZ1, that both genes fail to act epistatically in the control of plant growth (Castro et al. 2020). SUMO covalent attachment to a protein, known as sumoylation, normally occurs on a lysine within a consensus motif ΨKXE (Ψ, large hydrophobic residue; K, lysine; X, any amino acid; E, glutamic acid) of a target protein. Sumoylation follows a multi-step enzymatic cascade that includes deSUMOylating proteases (DSP)-dependent maturation of the preSUMO peptide, E1 activation, and E2 conjugation, normally facilitated by an E3 ligase. In addition, SUMO may also interact non-covalently with a protein, more specifically with a target’s SUMO-interacting motif (SIM, a hydrophobic amino acids stretch flanked by acidic residues) (Gareau and Lima, 2010). SUMO may control a target’s activity on multiple levels, including regulation of subcellular localization, modulation of protein interactions and complex assembly, changes in enzymatic activity, and potentiation/antagonization of other PTMs (Augustine and Vierstra, 2018).
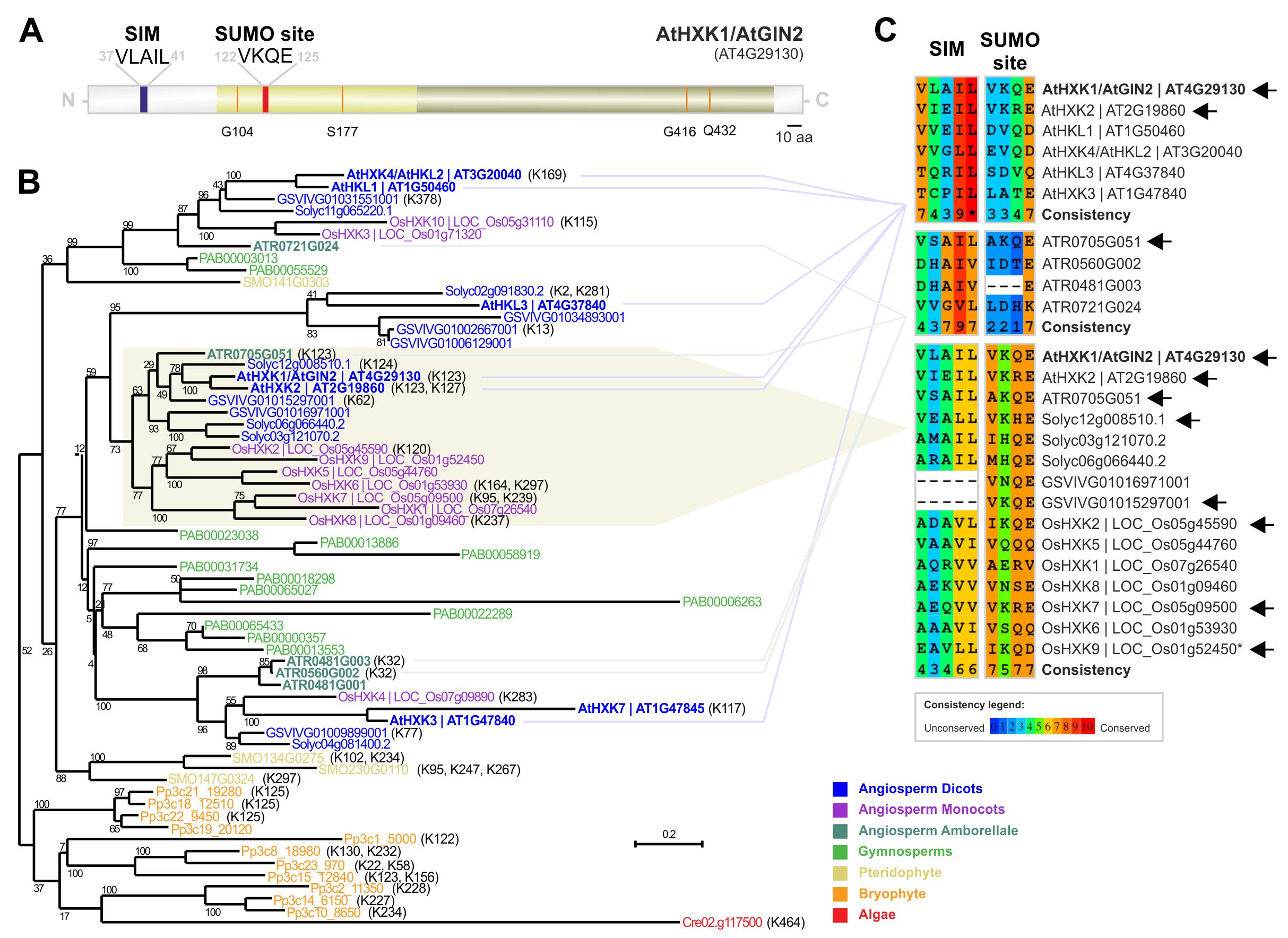
Here, we explore Arabidopsis HXK1 potential sumoylation sites, extended to other plant species to inspect the levels of evolutionary conservation. First, we submitted the Arabidopsis HXK1 protein sequence to the sumoylation site and SUMO-interaction site predictor software GPS-SUMO 1.0 (Zhao et al., 2014). This software predicted a high confidence sumoylation site in K123 and a SIM site (VLAIL motif) at position 37-41 (Fig. 1A). This prediction supports the hypothesis that sumoylation and/or SUMO interaction regulates HXK activity. Interestingly, the SUMO site is inside the predicted Hexokinase small subdomain, whereas the SIM is outside and closer to the N-terminus. To investigate if these motifs were conserved, indicating a conserved role for SUMO-HXK among plants, we performed a phylogenetic study in plant HXKs (Fig. 1B). HXK1 belongs to a multigene family that branched and expanded across plant evolution (Karve et al., 2010). While only one member exists in the algae Chlamydomonas reinhardtii, several HXKs are present in higher plants, ranging from four in the pteridophyte Selaginella moellendorffii, up to 13 members in the gymnosperm Picea abies (Fig. 1B). In Arabidopsis we observed the presence of seven members, one more than previous studies suggested (Karve et al., 2010). This additional protein, with AGI code AT1G47845, was probably overlooked in previous phylogenetic studies because of its truncated topology. None withstanding, the PFAM software predicted the existence of an HXK domain. In line with the previous AtHXKs naming order, we decided to designate AT1G47845 as AtHXK7.
Next we performed an alignment of Hexokinase members to inspect the level of conservation of AtHXK1’s K123 and SIM in all Arabidopsis paralogs (Fig. 1C, first panel). AtHXK7 was excluded from the analysis for being truncated in this region. Among Arabidopsis HXKs, only AtHXK1 (K123) and AtHKX2 (K123) revealed to have this lysine. AtHXK1 and AtHXK2 are members of the group 6 (Karve et al., 2010). Interestingly, the basal angiosperm Amborella trichopoda presented a member, ATR0705G051, clustered with AtHXK1/AtHXK2, that also contains a lysine (K123) with high/medium probability of sumoylation (Fig. 1C, second panel). In other Amborella trichopoda HXKs the lysine amenable to sumoylation exists but is in a different region of the protein (Fig. 1B,C). The fact that Amborella trichopoda protein ATR0705G051 clustered with dicots means that the Karve et al. (2010) classification into groups 6 and 7 (the latter only displaying monocots), should in fact be reconsidered as a single phylogenetic group. To evaluate if this lysine was conserved in group 6/7 we performed an alignment with all its members (Fig. 1C, third panel). Albeit not all HXKs within the group 6/7 contain the conserved motif, it is important to highlight that at least one HXK with the expected lysine exists in each species (Fig. 1C). In fact, the same lysine seems to be conserved across plant HXKs, being detected also in bryophytes and pteridophytes (Fig. 1B). The exception is the gymnosperm Picea abies, but this may derive from miss-annotation given the complexity of this genome. Results suggest that each species tends to retain at least one homologous K123 amenable to sumoylation during evolution.
The sumoylation of AtHXK1 was previously tested in bacteria, using an E. coli expressing a heterologous sumoylation system. AtHXK1 was positively sumoylated by both AtSUM1 and AtSUM3 isoforms (Elrouby and Coupland, 2010). However, the biological relevance of AtHXK1 sumoylation is yet to be determined. Nevertheless, we can discuss plausible scenarios. For instance, SUMO is involved in nuclear-cytoplasm trafficking and nuclear complex assembly (Palancade and Doye, 2008; Jentsch and Psakhye, 2013). The N-terminus of the human glucokinase (GK) is multi-sumoylated, which enhances glucose uptake, glycolysis and glycogen synthesis upon glucose treatment (Aukrust et al., 2013). Aukrust et al. (2013) proposed that SUMO contributes to GK activity by controlling the translocation between the cytoplasm and nucleus via the nucleoporin SUMO E3 ligase RANBP2. AtHXK1 is normally located in the mitochondria (Balasubramanian et al., 2007) but also, due to HXK1’s sugar signalling role, it can be relocated to the nucleus as part of a transcription repressor complex (Cho et al., 2006). In contrast, SUMO is majorly nuclear-located, but can also be detected in the cytoplasm (Lois et al., 2003; Saracco et al., 2007). It would be interesting to determine if SUMO can control AtHXK1 subcellular localization and if that has an impact on AtHXK1 activity. On the other hand, using GFP fusion constructs with OsHXK5 and OsHXK6, revealed that both retain a dual subcellular localization to mitochondria and the nucleus (Cho et al., 2009) and none of these contain the homologous K123. Another likely scenario is that SUMO may also interfere with protein-protein interactions or the assembly of protein complexes. A previous screening for HXK1-interacting partners identified the SNF1-related protein kinase (SnRK) regulatory subunit gamma 1 (SnRK1γ/KINγ) (Van Dingenen et al., 2019). SnRK1 is also a key regulator of cellular sugar and energy homeostasis of the cell (Crozet et al., 2014), and interestingly many of its subunits are modified and regulated by SUMO (Crozet et al., 2016). Increasing evidence points towards a strong SUMO-carbohydrate crosstalk (Castro et al., 2015; Castro et al., 2018b), and future efforts are necessary to uncover weather SUMO plays a role in HXK activity, and conversely determine the role of HXKs in SUMO-dependent carbohydrate signalling/metabolism regulation.
Methods
Request a detailed protocolThe AtHXK1/AtGIN2 sequence was retrieved from the database Plaza Dicots 4.0 (Van Bel et al., 2018) and submitted to Quick Scan mode of ScanProsite (de Castro et al., 2006) to scan for motifs/domains. To determine HXK sumoylation and SUMO-interaction sites, protein sequences were submitted for analysis in the software GPS-SUMO 1.0 (Zhao et al., 2014). The phylogeny of the plant Hexokinase gene family was carried out using the strategy previously described by Castro et al. (2018a). Amino acid sequences were retrieved using a combination of automated gene family assignment and curated homology analysis. More specifically, homologs of nine different species were retrieved from the comparative genomics database Plaza Dicots 4.0 (Van Bel et al., 2018), queried using the AGI code for AtHXK1/AtGIN2 (AT4G29130). Species were selected to include all major plant taxa (Bryophyte, Pteridophyte, Gymnosperms, the basal Angiosperm Amborella trichopoda, Angiosperm Monocots and Angiosperm Eudicots including the model Arabidopsis thaliana). The ancestral Algae species, Chlamydomonas reinhardtii, was used as outgroup. Sequences indicated by the Plaza database as outliers were excluded from the analysis. For all protein sequences, we detected the presence of Pfam protein domains in a batch search using HMMSCAN (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) and confirmed them to have an hexokinase domain. Amino acid sequences were aligned using MAFFT (v7.402) on CIPRES (Katoh and Standley, 2013). We tested for the substitution best-fit model using ProtTest v3.3 (Darriba et al., 2011). The phylogenetic tree was estimated using maximum likelihood (RaxML v8.2.12 on CIPRES) with a JTT substitution model and 1000 bootstrap iterations. Computation was run at the CIPRES Science Gateway V3.3 (http://www.phylo.org) (Miller et al., 2011). The tree was visualized using SeaView v4.4.0 (Gouy et al., 2010). Sequence alignments for specific sets of proteins were conducted using PRALINE (Simossis and Heringa, 2005). To determine gene nomenclature/designation, Arabidopsis and rice gene IDs were submitted to _at to AGI Conversion Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_agi_converter.cgi) and Rice Genome Database Project (http://rice.plantbiology.msu.edu/), respectively.
References
Funding
Financial support was provided by Fundação para a Ciência e Tecnologia (FCT/MCTES) to H.A. [CEECIND/00399/2017/CP1423/CT0004] and S.F. [SFRH/BD/120020/2016]. Also from FCT/MCTES, Fundo Europeu de Desenvolvimento Regional (FEDER), and COMPETE – POCI – Programa Operacional Competividade e Internacionalização, to P.H.C. [PTDC/BAA-AGR/31122/2017, POCI-01-0145-FEDER- 031122].
Reviewed By
AnonymousHistory
Received: January 30, 2020Revision received: April 2, 2020
Accepted: May 29, 2020
Published: June 29, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Castro, PH; Freitas, S; Azevedo, H (2020). Plant hexokinase phylogenetic analysis highlights a possible regulation by the posttranslational modifier SUMO. microPublication Biology. 10.17912/micropub.biology.000260.Download: RIS BibTeX