Department of Biochemistry and Molecular Biology, The Pennsylvania State University
Description
Caenorhabditis elegans uv1 cells undergo a dramatic cell death in a pnc-1 NAD+ salvage deficient mutant (Huang and Hanna-Rose, 2006). uv1 cell death is a result of accumulation of the PNC-1 substrate nicotinamide (NAM), which overactivates the OCR-4 OSM-9 transient receptor potential cation V (TRPV) channel, causing excitotoxicity (Upadhyay, et al., 2016, Vrablik, et al., 2009). Reduced uv1 cell survival is almost completely restored in pnc-1 mutant animals by sustained activation of the Epidermal Growth Factor (EGF) receptor LET-23, either by overexpression of the ligand LIN-3 or by a gain of function mutation in the receptor (Huang and Hanna-Rose, 2006). We presume that the constitutively activated receptor acts cell autonomously in the uv1 cells to promote survival, but have no direct evidence in support of that presumption, besides a prior report showing that a let-23(sa62) gain of function allele was able to induce uv1 specification in the absence of LIN-3 (Chang, et al., 1999). To find genes required for uv1 survival in a let-23(sa62gf); pnc-1(pk9605) background, we performed an RNAi screen and found that phosphatidylcholine synthesis via PMT-1 or PCYT-1 is required for LET-23 to promote uv1 cell survival in pnc-1 background (Crook, et al., 2016). Moreover, treatment with exogenous phosphatidylcholine alone is partially sufficient to promote cell survival in the pnc-1 mutant (Crook, et al., 2016). The requirement of phosphatidylcholine synthesis indicates that membrane phospholipid composition, and by extension disruption of lipid homeostasis, may play a role in preventing TRPV-induced excitotoxic death.
Lipid synthesis homeostasis in general and phosphatidylcholine synthesis in particular are regulated by the srebp-1 homolog sbp-1 in C. elegans (Walker, et al., 2011). SBP-1 is activated by low phosphatidylcholine levels resulting from pmt-1 or pcyt-1 RNAi (Walker, et al., 2011). We hypothesized that uv1 survival in a let-23gf: pnc-1 background decreases when SBP-1 is activated through reduced phosphatidylcholine synthesis via pmt-1 or pcyt-1 RNAi. We predicted that inactivation of SBP-1 when pmt-1 or pcyt-1 were knocked down by RNAi would restore uv1 survival to that seen in a let-23gf: pnc-1 background.
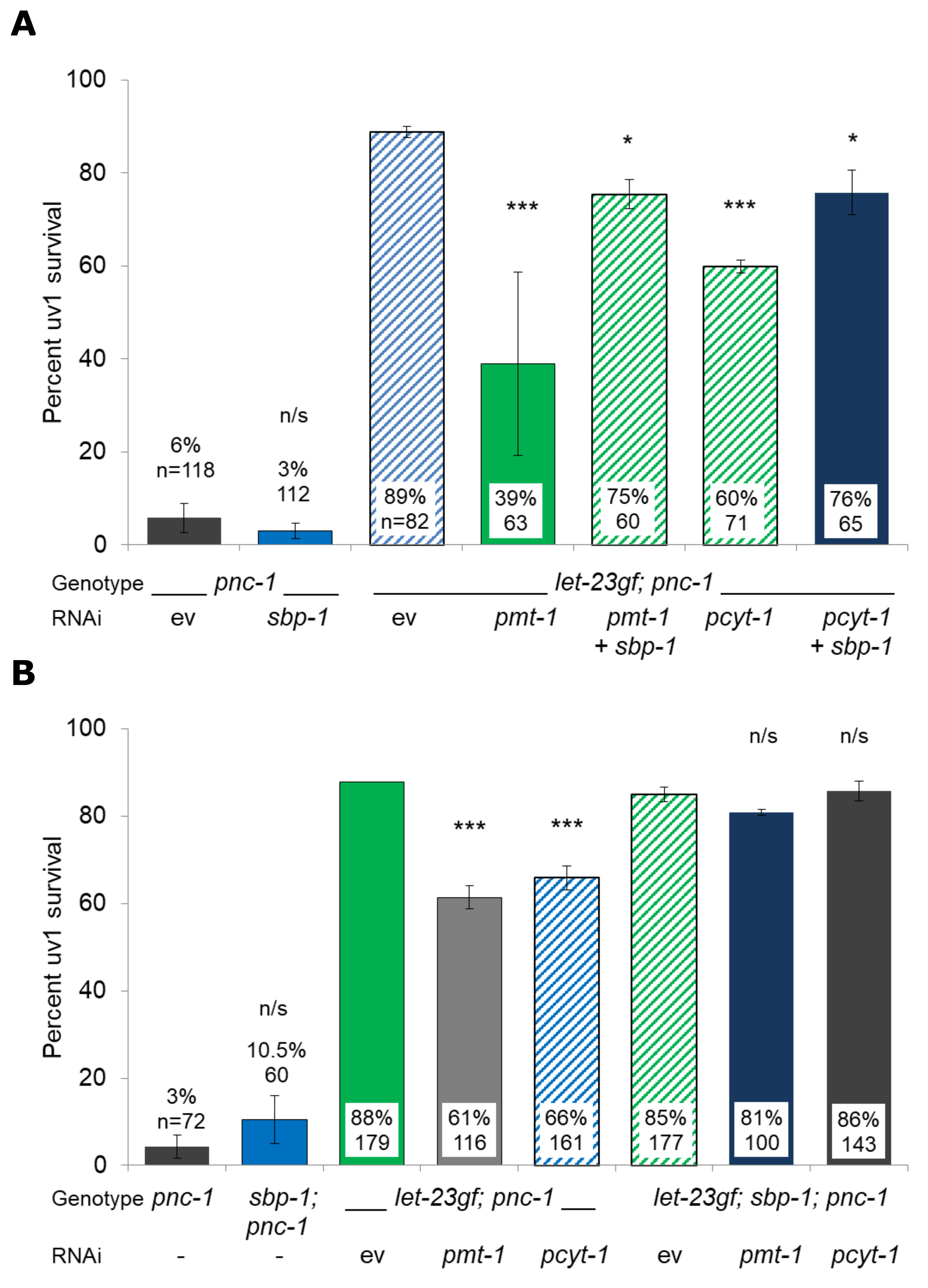
We tested the role of SBP-1 by reducing its activity in a let-23(sa62gf); pnc-1(pk9605) background with reduced phosphatidylcholine synthesis. We took two complementary approaches. First we carried out double RNAi experiments (Ahringer, 2006) where both sbp-1 and pmt-1 or pcyt-1 were knocked down in a let-23(ga62gf); pnc-1(pk9605) background using an extrachromosomal ida-1p::gfp marker. Each RNAi was carried out twice with an empty vector (L4440) as control. Double RNAi experiments were carried out by mixing equal volumes of overnight LB-Amp RNAi cultures before spotting NGM-Carb-Tet plates. We found that double RNAi of sbp-1 with either pmt-1 or pcyt-1 resulted in a significant increase in uv1 survival compared with pmt-1 or pcyt-1 RNAi alone (Fig. 1A). However, uv1 survival was not restored to the level seen in a let-23(sa62gf); pnc-1(pk9605) background, which may have been due to incomplete knockdown in the double RNAi experiments. To address this possibility, we created a let-23(ga62gf); sbp-1(ep79); pnc-1(pk9605) strain with an integrated ida-1p::gfp marker. We then knocked down pmt-1 or pcyt-1 in this background and compared its effect on uv1 survival to the same gene knockdowns in a let-23(ga62gf); pnc-1(pk9605) strain, using an integrated ida-1p::gfp marker to count living uv1 cells. Each RNAi was carried out three times. We found that the pmt-1 or pcyt-1 knockdown induced reduction of uv1 survival in a let-23(ga62gf); pnc-1(pk9605) background did not occur when these genes were knocked down in a let-23(ga62gf); sbp-1(ep79); pnc-1(pk9605) background (Fig. 1B). There was no effect of either sbp-1 RNAi (Fig. 1A) or the sbp-1(ep79) mutant allele (Fig. 1B) alone on uv1 cell survival in a pnc-1 mutant background. Thus, both of our approaches support the hypothesis that sbp-1 is required for the reduction in uv1 survival seen when phosphatidylcholine synthesis is reduced in a let-23(ga62gf); pnc-1(pk9605) background.
let-23(sa62gf) mediated uv1 survival requires phosphatidylcholine synthesis and reducing phosphatidylcholine synthesis reduces cell survival. Our work shows that the reduction in uv1 survival when pcyt-1 or pmt-1 are knocked down is dependent on SBP-1. We propose the following model based on these results and our previous work on TRPV-induced excitotoxic death (Upadhyay, et al., 2016). An elevated level of NAM in a pnc-1 mutant activates the OCR-4/OSM-9 TRPV channel and results in uv1 cell death. Constitutive activation of LET-23 promotes phosphatidylcholine synthesis via PMT-1 and PCYT-1. Elevated phosphatidylcholine levels result in a cell membrane in which the OCR-4/OSM-9 TRPV channel is nonfunctional, which in turn prevents its activation by elevated NAM levels and results in uv1 survival. Reduction of phosphatidylcholine levels via pmt-1 or pcyt-1 RNAi activates SBP-1, which restores lipid homeostasis and cell membrane phospholipid composition, resulting in a functional OCR-4/OSM-9 TRPV channel and cell death. Support for our model also comes from the observation that exogenous phosphatidylcholine restores OLQ survival in a pnc-1 background, even though constitutive activation of LET-23 did not (Crook, et al., 2016). The precise mechanism by which elevated phosphatidylcholine levels disrupt OCR-4/OSM-9 TRPV channel function is not yet known, but our work supports an interaction between phosphatidylcholine levels, ion channel function and SBP-1-mediated lipid homeostasis.
Reagents
Strains:
HV560 inIs179[ida-1p::gfp] II; pnc-1(pk9605) IV
HV692 let-23(sa62)gf II; pnc-1(pk9605) IV; Ex[Pida-1p::gfp 7.6]
HV776 let-23(sa62)gf inIs179[ida-1p::gfp] II; pnc-1(pk9605) IV
HV790 inIs179[ida-1p::gfp] II; sbp-1(ep79) III; pnc-1(pk9605) IV
HV792 let-23(sa62)gf inIs179[ida-1p::gfp] II; sbp-1(ep79) III; pnc-1(pk9605) IV
Ex[Pida-1p::gfp 7.6] is from Zahn et al., 2001. None of these strains are or will be available at the CGC. The strains used in this study are available from the authors upon request.
We used the following clones from the Ahringer RNAi library: pmt-1 ZK622.3 II-4G04, pcyt-1 F08C6.2 X-3N20, and sbp-1 Y47D3B.7 III-6C01.
Acknowledgments
Some strains used to make the strains in this study were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
Funding
This work was funded by NIH grant R01GM086786 to WHR
Reviewed By
Anonymous, Bhagwati Gupta and Chieh ChangHistory
Received: January 30, 2020Revision received: February 12, 2020
Accepted: June 1, 1970
Published: June 29, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Crook, M; Hanna-Rose, W (2020). Overactive EGF signaling promotes uv1 cell survival via increased phosphatidylcholine levels and suppression of SBP-1. microPublication Biology. 10.17912/micropub.biology.000266.Download: RIS BibTeX