University of Utah Bioscience Program, Salt Lake City, UT
Description
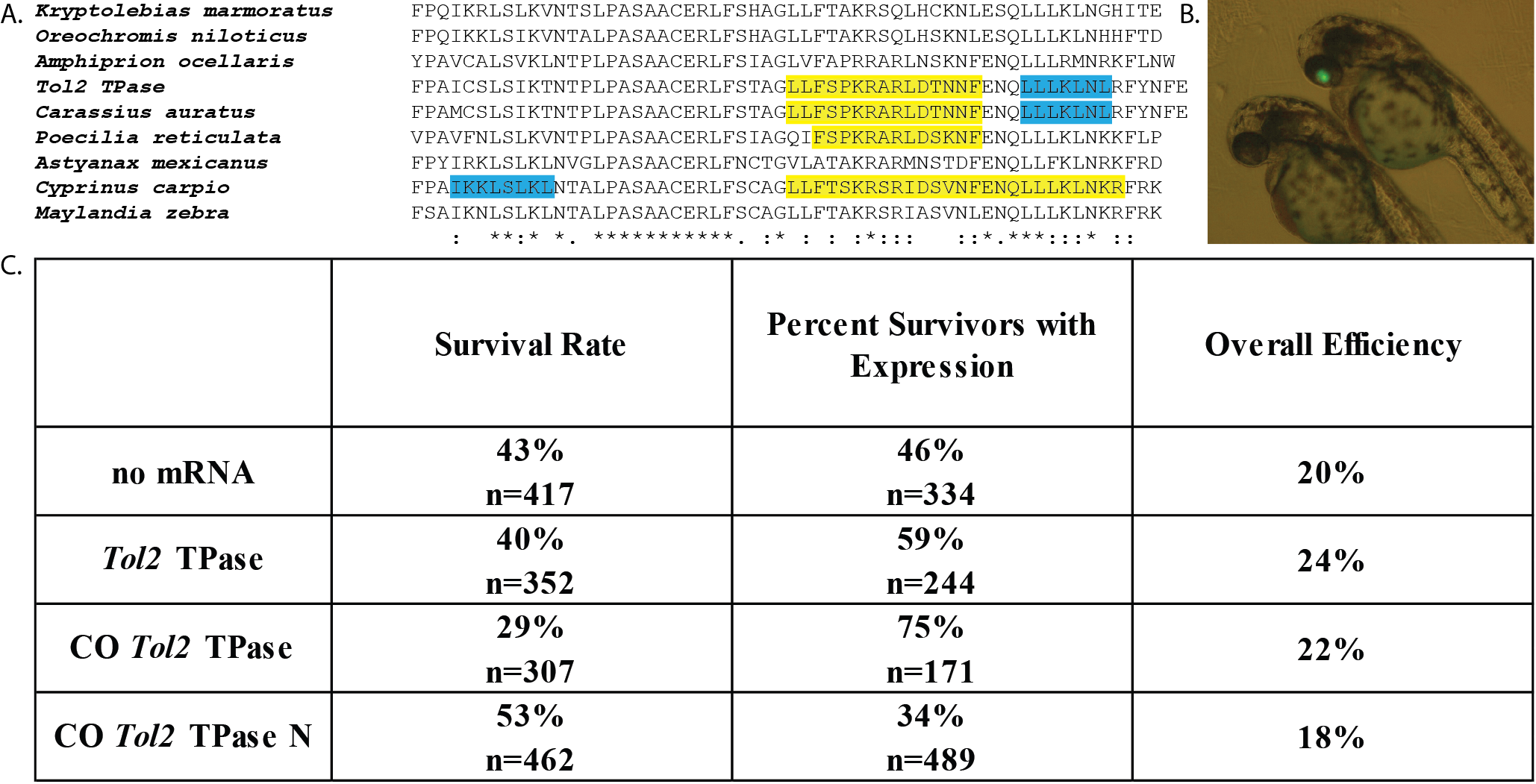
Type II transposable elements (TEs) are segments of DNA that can be mobilized within the genome through the action of transposase (TPase) proteins (Craig 2002). In general, the rate at which TPase proteins bind the terminal sequences of the elements to form a functional transposition complex determines the relative mobility of the element (Mizuuchi et al. 1992; Zayed et al. 2004; Zhang et al. 2001). The rate of transposition complex formation is thus determined by the concentration of functional TPase protein, the number of TE sequences present, and the localization of the TPase proteins within the cell. Several studies suggest that transposition complex formation is regulated by access to the nucleus, as alteration of nuclear localization signals (NLS) and nuclear export signals (NES) influences transposition (Hancock et al. 2010; Payero et al. 2016; Ramakrishnan et al. 2019). Zebrafish studies often involve the integration of DNA sequences (i.e. overexpression cassettes) into embryos, leading to transient or germ line expression (Nusslein-Volhard and Dahm 2002). Inserting transgenes between TE sequences and providing a TPase protein source has been shown to increase transgenesis efficiency across multiple model organisms (Ding et al. 2005; Ivics and Izsvák 2004; Munoz-Lopez and Garcia-Perez 2010; Zayed et al. 2004). The Tol2 TE from Medaka fish (Oryzias latipes) has been developed as a means to improve zebrafish transgenesis, including both transient and germline integration (Kawakami 2007; Koga et al. 1996; Kwan et al. 2007; Ni et al. 2016). Tol2-mediated transgenesis is induced by co-injecting Tol2 TPase mRNA together with a Tol2 terminal sequence-flanked construct into 1-4 cell stage zebrafish embryos (Kawakami 2007; Kwan et al. 2007; Ni et al. 2016). Previous reports indicated that addition of the Tol2 TPase mRNA (expressed from pCS2FA) produced about a 3-fold increase in Tol2 flanked transgene expression compared to control (Kwan et al. 2007). Another study demonstrated that a zebrafish codon optimized version of Tol2 TPase led to successful germline transmission, but no direct comparison of efficiency was reported (Suster et al. 2011). The goal of this study was to explore the extent that efficiency of Tol2-mediated transgene expression could be improved by codon-optimizing the Tol2 TPase gene for zebrafish or altering a detected NES and NLS in the Tol2 TPase (Figure 1A). We hypothesized that increasing the translation efficiency (Gustafsson et al. 2004) and access to the nucleus would potentially improve transposition complex formation in transience. Analysis of the Tol2 TPase codon usage showed that it contained 19 codons (TTA, CTA, and TCG) that are rarely used by zebrafish (Nakamura et al. 2000). Codon optimization resulted in the development of a CO Tol2 TPase construct that more appropriately matched the zebrafish-specific codon bias, without altering the amino acid sequence (Zhou et al. 2016). In the CO Tol2 TPase N construct, the NES in the Tol2 TPase (Figure 1A) was removed by changing L641A and L643A and the existing NLS (PKRARLD, NLS score=8.5) was strengthened by changing it to PKKKRKV (NLS score=13) (Dingwall and Laskey 1991; Kosugi et al. 2009).
Injection of 1-4 cell-stage zebrafish embryos with pDestTol2pACryGFP showed a significantly increased frequency of eGFP expression in the eye (i.e. Figure 1B) when co-injected with wild type Tol2 TPase mRNA compared to no mRNA (Figure 1C, p=0.0024). This is below the reported 3-fold increased frequency of expression previously reported using non-codon optimized Tol2 (Kwan et al. 2007). We observed a significantly increased frequency of eye eGFP expression when we used the codon optimized (CO Tol2 TPase) mRNA (Figure 1C, vs. no mRNA p=<0.0001, vs. WT TPase p=0.0008). These results suggest that the codon optimized version of the mRNA is translated more effectively in embryos, leading to an overall higher concentration of Tol2 TPase protein. In contrast, the NES and NLS altered version (CO Tol2 TPase N) showed decreased expression frequency compared to all other groups (vs. no mRNA p=0.0013; vs. WT Tol2 TPase p<0.0001; vs. CO Tol2 TPase p<0.0001). This suggests that the changes made to the NES and NLS may have disrupted overall protein function of the CO Tol2 TPase N protein (Figure 1C).
When both the expression frequencies and survival rates were used to calculate overall transgene expression efficiency, we observed that all treatments resulted in about 20% of the injected eggs producing eGFP-expressing embryos surviving to 2 dpf (Figure 1C). An inverse relationship was observed between the survival frequency and the frequency of eGFP expression at 2 dpf. We observed that significantly less fish developed properly with the higher activity CO Tol2 TPase mRNA compared to no mRNA (p = 0.0002) and the WT Tol2 TPase mRNA (p=0.0033) (Figure 1C). This suggests that genome disruption potentially caused by transgene insertion may be an important limit on transgenesis rates.
The high frequency of eGFP expression observed in this study for the no mRNA control suggests that a high percentage of fluorescence observed was not TPase mediated. In addition, our results suggest that the addition of Tol2 mRNA does not drastically increase the overall rate of transient expression of transgenes (total number of surviving transgenic fish/ embryos injected). However, there are significant benefits to the Tol2 system, as a greater fraction of fish that survive show transient transgene expression, potentially leading to easier screening for expressing embryos. Our results suggest that co-injecting constructs with the CO Tol2 TPase mRNA (available through Addgene #133032) is a significant improvement over the existing technology. This codon optimized Tol2 has the potential to increase the yield of transient expression and should be tested for its ability to induce heritable transgenesis. Although this study did not examine heritable transgenesis, the next step in this investigation would be to examine the rates of integration of the pDestTol2pACryGFP transgene in fish injected with each mRNA.
Methods
Request a detailed protocolSequence analysis
The Tol2 TPase sequence (GenBank: BAA87039.1) was used in NCBI’s Basic Local Alignment Search Tool (BLAST) to obtain homologs. The alignment was performed using EMBL-EBI Clustal Omega Multiple Sequence Alignment tool (Sievers et al. 2011). NLS and NES sequences were identified using cNLS Mapper (Kosugi et al. 2009) and NetNES 1.1 Server (La Cour et al. 2004), respectively.
Construction of novel plasmids
IDT’s codon optimization tool (https://www.idtdna.com/codonopt) was used to generate a novel sequence that altered most of the codons, removing TTA, CTA, and TCG codons. The two new versions of Tol2 TPase were synthesized as IDT gBlocks®, cloned into the BamHI and XbaI sites of pCS2FA plasmid (Kwan et al. 2007), and sequence verified. Plasmid DNA was purified using Zyppy™ Plasmid Miniprep and quantitated by Qubit® dsDNA BR Assay.
mRNA preparation
Plasmids were linearized with NotI-HF, column purified using the Zymo DNA Clean and Concentrator™ -5 kit and quantitated by Qubit® dsDNA BR Assay. In vitro transcription was performed on 1 µg of DNA using the Ambion mMessage mMachine™ SP6 Kit. After the transcription reaction, the product was treated with TURBO DNase and cleaned using the Nucleospin® RNA II columns. Gel electrophoresis was used to confirm for the presence of mRNA and the concentration was determined by Qubit® RNA BR Assay.
Injection of mRNA and screening for GFP expression in zebrafish
A previously established transgenesis assay (Berger and Currie 2013) in which pDestTol2pACryGFP produces eye specific eGFP expression from the crystallin (cryaa) promoter (Kurita et al. 2003) was used. pDestTol2pACryGFP was a gift from Joachim Berger & Peter Currie (Addgene plasmid # 64022 ; http://n2t.net/addgene:64022 ; RRID:Addgene_64022). eGFP fluorescence was detected by fluorescence microscopy 2 dpf (Figure 1B). Adult wild-type AB (ZFIN) zebrafish were mated and injections were performed on resulting embryos within one hour post fertilization (1-4 cell stage). Each embryo was injected with 25 pg of mRNA and 30 pg of pDestTol2pACryGFP DNA as described (Kwan et al. 2007). Two treatments were injected each day until 6 replicate injections of each mRNA were achieved. The order of treatments was continually altered to prevent injection timing bias. Screening of the embryos took place at 2 dpf on either an Olympus CKX41 Inverted Microscope or an Olympus SZX12 Fluorescence Stereo Microscope.
Statistical Analysis
eGFP expression and survival data were pooled and analyzed using a contingency table. A χ2 analysis was used to identify significance and multiple comparisons were conducted using Fisher’s exact test (SAS version 9.4). The null hypothesis was that the proportion observed was independent of the treatment.
Acknowledgments
Special thanks to Dr. Derek Zelmer (USCA) for assistance with statistical analysis.
References
Funding
This work was supported by P20GM103499 (SC INBRE) from the National Institute of General Medical Sciences, National Institutes of Health and NSF grant #1651666. This work was partially supported by the University of South Carolina Magellan Scholar program to Allison S. Mackey.
Reviewed By
Anonymous and Bruce DraperHistory
Received: April 13, 2020Revision received: June 8, 2020
Accepted: June 11, 2020
Published: June 11, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Mackey, AS; Redd, PS; DeLaurier, A; Hancock, CN (2020). Codon optimized Tol2 transposase results in increased transient expression of a crystallin-GFP transgene in zebrafish. microPublication Biology. 10.17912/micropub.biology.000268.Download: RIS BibTeX