Description
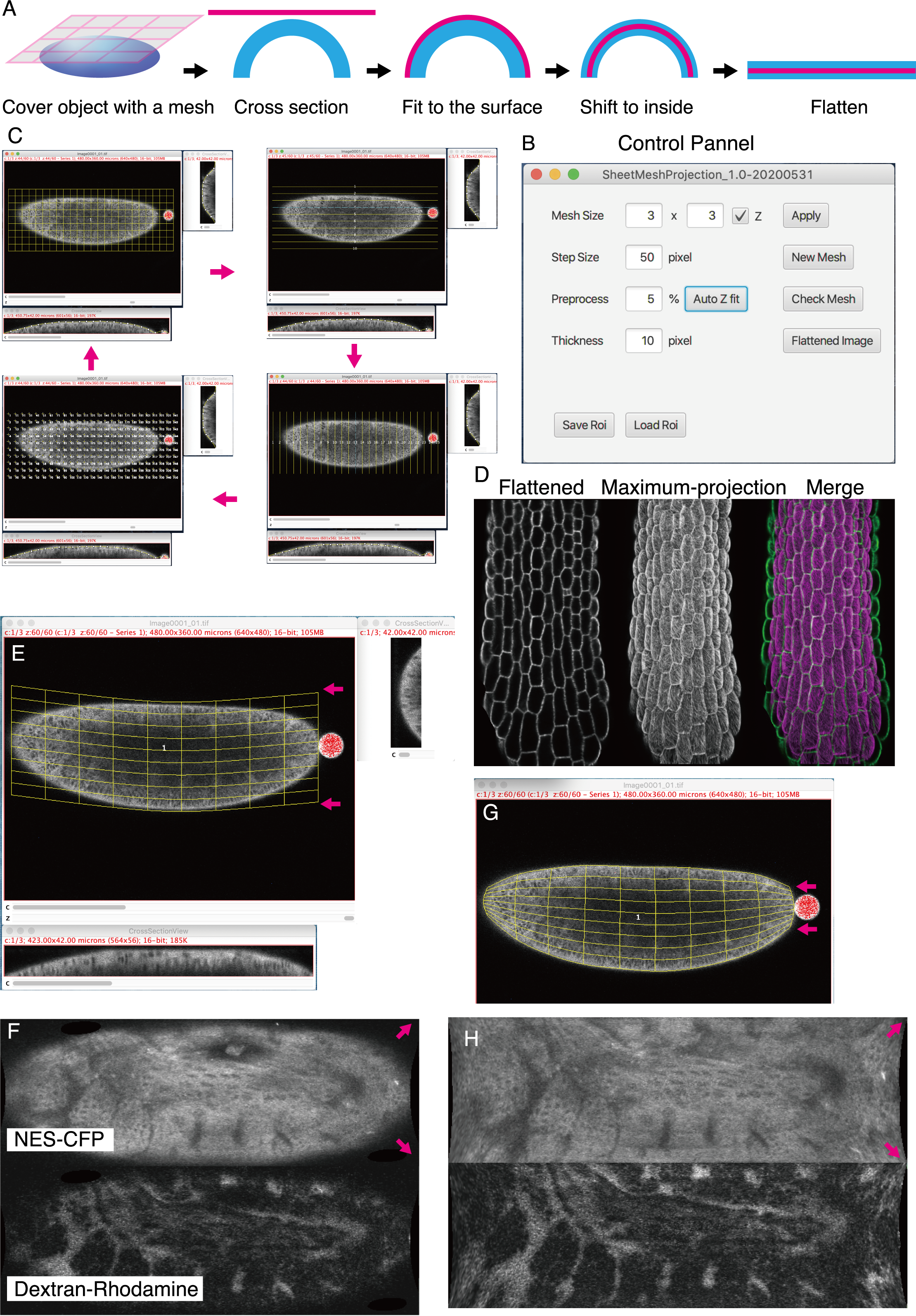
Optical cross-sectioning of whole embryos with advanced fluorescent microscopes such as confocal laser scanning microscopes or light-sheet microscopes has been extensively used in developmental biology. The maximum projection of markers localized to the object surface is widely used to reconstruct surface profiles (Erguvan et al., 2019). However, this technique is effective only to the surface parallel to the plane of sections, and area profiles of diagonal surfaces, such as the margin of embryos, are under-represented. Tools for tracing curved surfaces have been developed (de Reuille et al., 2015; Heemskerk & Streichan, 2015; Viktorinová et al., 2019), but their usage is not easy for novice users. A simple and easy-to-use tool with basic surface extraction function for users of the most widely used image analysis platform, ImageJ (https://imagej.nih.gov) is anticipated. For this reason, we developed a highly versatile ImageJ plugin “SheetMeshProjection” for extracting curved surfaces with user-defined thickness and creating expanded 2D images.
A step-by-step manual is provided in the plugin package. Unlike conventional projection software that recognizes the surfaces pixel by pixel, SheetMeshProjection uses a coarsely spaced mesh to capture the surfaces of objects like casting a net (Fig. 1A). If necessary, each line of the mesh is manually corrected to fit the surfaces in x-z or y-z cross-sectional view, and lines can be adjusted to longitude to cover spherical objects (Fig. 1G, H). Mesh line position can be moved to avoid positions of hairs and other small appendages that potentially confuse surface tracking. Mesh density can be increased later to refine tracking (Fig. 1B). After fitting the mesh line position, the surfaces of user-defined thickness is extracted and converted to a flattened object, like skinned fur rugs.
Images extracted in this way provide more faithful representations of the surface of 3D objects with acute curvature, especially at the margin (Fig. 1D). It should be noted that the appearance of flattened images varies if location and spacing of mesh lines are changed (Fig. 1E-H) and quantitative information (length and area) of the extended surfaces may not be preserved. Flattened images should be regarded as qualitative representations of the object surfaces. For the precise acquisition of 2D-projected surface images, more sophisticated software is available (Schmid et al., 2013), although it is limited to spherical objects. Flattened surface images of whole embryos or organs with convex surfaces may be used to help analysis of cell counting and cell neighborhood analysis and should help studies of complex cell composition of 3D biological objects.
Reagents
Software Availability: The plugin is available at Fiji update site (https://sites.imagej.net/SheetMeshProjection/). Full package of plugins, instruction manual, and a test image is available from the authors’ home page https://signaling.riken.jp/en/en-tools/imagej/1743/). Source code and documentation (ver. 1.0-20200721, MIT) are available on GitHub (https://github.com/Wada-H/SheetMeshProjection). An archived version of this data is available on Caltech Data: DOI 10.22002/D1.1606.
Drosophila strain: CFP fluorescence of the pubi-ekarev-nes58A FRET sensor strain (Ogura et. al., 2018) was shown as the NES-CFP image (Fig. 1F).
Acknowledgments
We are grateful to Zhengkuan Sun and Wei-Chen Chu in Hayashi lab for comments. We thank Kentaro Yoshida (RIKEN) and Stéphane Verger (Swedish University of Agricultural Sciences) for sharing image data.
References
Funding
Grant in Aid for Scientific Research from MEXT Japan 19H00996.
Reviewed By
Steven Marygold and Pavel TomancakHistory
Received: April 30, 2020Revision received: August 17, 2020
Accepted: August 19, 2020
Published: August 19, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Wada, H; Hayashi, S (2020). Net, skin and flatten, ImageJ plugin tool for extracting surface profiles from curved 3D objects. microPublication Biology. 10.17912/micropub.biology.000292.Download: RIS BibTeX