University of Warwick, UK
Description
UPF1 is an RNA helicase that scans RNA to unwind secondary structures and to displace associated factors (Franks et al., 2010; Fiorini et al., 2015; Lee et al., 2015; Kanaan et al., 2018). UPF1 has been mostly studied for its role in nonsense mediated mRNA decay (NMD) and other translation-dependent RNA quality-control pathways in the cytoplasm (Isken and Maquat, 2008; Kim and Maquat, 2019). However, we have recently reported that UPF1 associates, genome wide, with nascent transcripts at most Pol II transcription sites in Drosophila (Singh et al., 2019). The association of UPF1 with nascent transcripts appears to be necessary for the release of processed polyadenylated mRNAs from their transcription sites and also for their export from nucleus to cytoplasm (Singh et al., 2019).
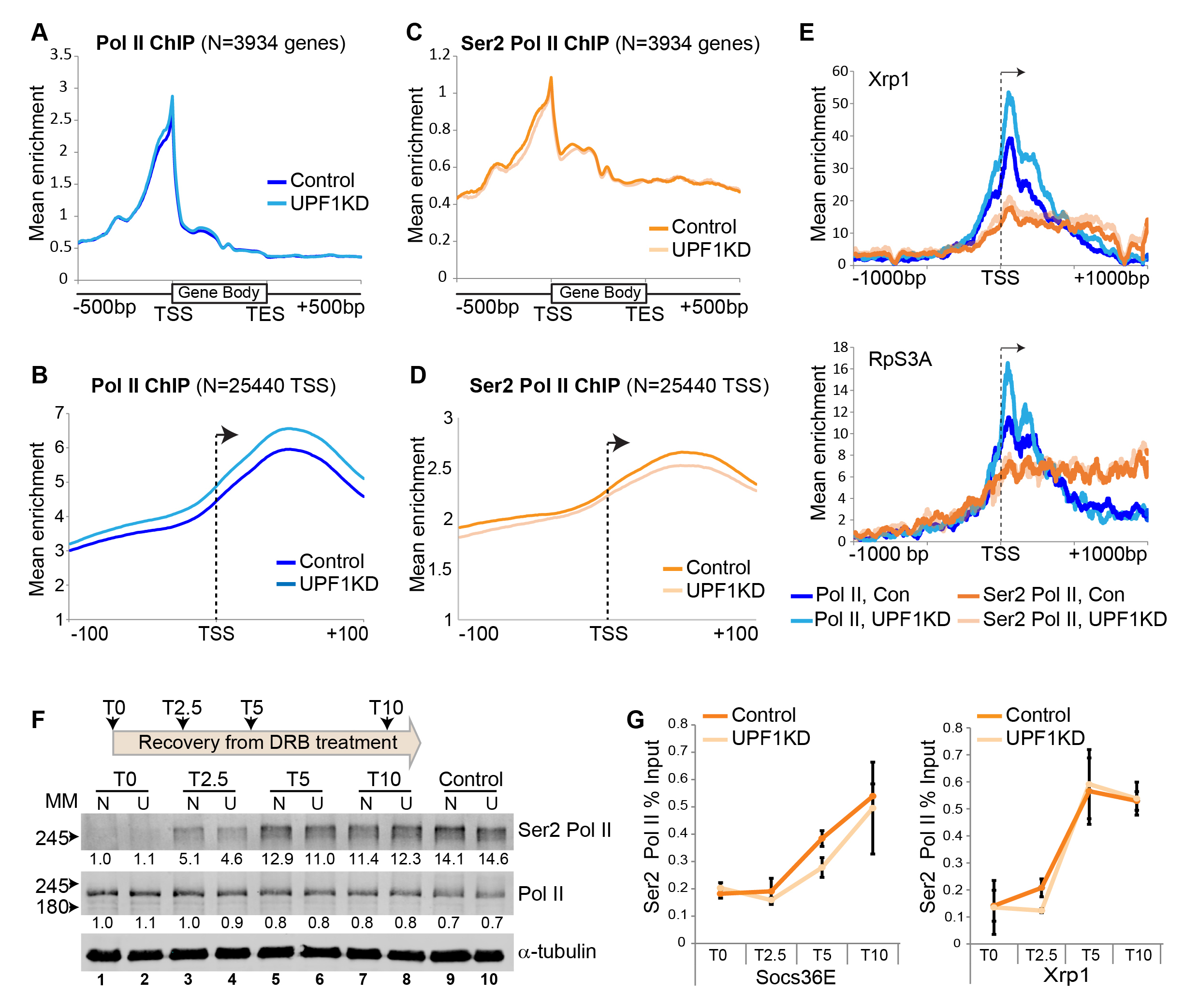
Here we considered whether the association of UPF1 with nascent transcripts influences Pol II transcription, and tested it by ChIP-seq of Pol II in Drosophila S2 cells. We discovered (using two biological replicates) that there is significantly more unphosphorylated Pol II associated with the start of genes in cells depleted of UPF1 (Figure 1A and 1B). This difference is most apparent 20 to 60 base pair (bp) downstream of the transcription start site (TSS), as shown in the expanded depiction in Figure 1B which is based on many more individual TSSs. This is measured by quantifying the aggregate Pol II signal over a +/- 100 bp span at each of 25440 TSSs (P = 0.011). A 1.2-fold or greater increase of unphosphorylated Pol II in UPF1-depleted cells was detected at 1990 TSSs. The 20-60 bp downstream region corresponds to TSS-proximal Pol II pausing sites in Drosophila (Adelman and Lis, 2012). Conversely, the amount of Ser2 Pol II associated with these TSSs was unchanged or marginally reduced in UPF1-depleted cells (Figure 1C and 1D). The increase in unphosphorylated Pol II loading downstream of the TSS in UPF1-depleted cells, alongside fairly constant Ser2 Pol II loading, is illustrated here by Xrp1 and RpS3A (Figure 1E), two genes that are highly transcribed and show strong UPF1 association in normal cells (Singh et al., 2019).
We considered whether this increase in unphosphorylated Pol II at pausing sites might be a consequence of reduced Ser2 phosphorylation, resulting in the slow release of Pol II from pausing sites and abnormal transcription elongation. We tested this hypothesis by examining the reappearance of newly phosphorylated Ser2 Pol II following withdrawal of 5, 6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB), which is a transcription inhibitor that blocks Ser2 phosphorylation and should lead to rapid depletion of Ser2 Pol II. As expected, DRB treatment of S2 cells causes a drastic depletion of Ser2 Pol II and an increase of unphosphorylated Pol II (Figure 1F, compare control lanes 9 and 10 with DRB-treated lanes 1 and 2). Ser2 Pol II levels began to recover soon after DRB removal and were similar to those of untreated control cells within 10 minutes (Figure 1F, compare lanes 7-8 with control lanes 9-10). However, the recovery in UPF1-depleted cells seemed slower during the first 2.5 minutes and to a lesser extent after 5 minutes (Figure 1F, compare lanes 3 vs. 4 and 5 vs. 6). Similar observations were made in another independent replicate of the experiment. A comparable blunted recovery of gene-associated Ser2 Pol II in UPF1-depleted cells was detected by ChIP-PCR at the two representative highly active genes, Socs36E and Xrp1 (Figure 1G), both of which show strong association with UPF1 (Singh et al., 2019). RpS3A was not examined because it is too short (about 2 kb) for this assay.
These observations indicate that UPF1 might, by associating with nascent transcripts, influence Ser2 phosphorylation of Pol II at TSS-proximal pausing sites and hence transcription elongation at several Drosophila genes. Alternatively, or additionally, UPF1 might affect premature transcription termination and Pol II release from DNA downstream of the TSS, which are other, perhaps more important determinants of the high accumulation of Pol II at the beginning of genes (Kamieniarz-Gdula and Proudfoot, 2019) .
Methods
Request a detailed protocolCell culture and RNA interference
S2 cells (CVCL_Z232, laboratory stock originally purchased from Invitrogen/Thermo Fisher Scientific) were cultured in Insect–XPRESS media (Lonza) supplemented with 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin-Glutamine mix (P/S/G, Invitrogen) at 27°C. To make the RNAi constructs for UPF1, the specific sequences were PCR amplified from S2 cell genomic DNA by using corresponding primer pairs. Along with the desired gene sequence, the primer pair carried the T7 promoter sequence at their 5’ end. The amplified PCR fragments were purified using Monarch® PCR and DNA Cleanup Kit (T1030S, NEB) and dsRNA was synthesized using the T7 RiboMAX express RNAi system (P1700, Promega). To induce RNAi, a six-well culture dish was seeded with 106cells/well in serum-free media and mixed with 15 µg of dsRNA/well. Following 1 hour incubation at room temperature, 2 mL of complete media was added to each well and the cells were incubated for the next three days to knockdown UPF1 before harvesting.
Western blotting and antibodies
Western blotting was performed as described previously (Singh et al., 2019). The antibodies used in western blotting were diluted as follow: Rabbit anti-Ser2 Pol II (Abcam, ab5095, 1:5000), Mouse anti-Pol II (Abcam, ab817 (8WG16), 1:5000) and mouse anti-β-tubulin (Sigma-Aldrich, T5168 1:5000).
DRB treatment
S2 cells were treated with DRB (Sigma-Aldrich, 125µM) for 1 hour at room temperature. Ser2-Pol II ChIP was performed as described below and quantified by real-time PCR using Socs36E and Xrp1 primers shown below.
ChIP-seq and data analysis
Chromatin immunoprecipitation, sequencing and data analysis was done as described previously (Singh et al., 2019). For each ChIP, typically we used 5 µg of antibody including Rabbit anti-Ser2 Pol II (Abcam, ab5095) and Mouse anti-Pol II (8WG16, Abcam, ab817). ChIP-signal quantification and metagene analysis were performed as previously described (Singh et al., 2019). All new ChIP-seq raw sequencing FASTQ data and Bedgraph files were deposited in the GEO repository (Accession No GSE157612), all other datasets used in this study were previously deposited (Accession No GSE16808).
Primers List:
UPF1RNAi-FP – 5’-TTAATACGACTCACTATAGGGGAGAGGAGAAGCCAGGCATTGA-3’
UPF1RNAi-RP – 5’-TTAATACGACTCACTATAGGGGAGAGACCGTGGCCCAACAGG-3’
(Bold is T7 promoter sequence)
Socs36E-FP – 5’-CAGAAAACCGCACACAGACA -3’
Socs36E-RP – 5’-CACACATCGGACTAACAGCG-3’
Xrp1-FP – 5’-TCATAATGCTTGTGGGGCCT -3’
Xrp1-RP – 5’-AGGGTCCCTCTAAACAAGCT-3’
Reagents
See Methods
Acknowledgments
We like to thank Hannah L Dixon and Bob Michell for critically reading the manuscript.
References
Funding
Leverhulme Trust, RPG-2014-291, Saverio Brogna; Wellcome Trust, 9340/Z/09/Z, Saverio Brogna; BBSRC, BB/M022757/1, Saverio Brogna; BBSRC , BB/S017984/1, Saverio Brogna; BBSRC, BB/M017982/1, Daniel Hebenstreit; BBSRC, BB/L006340/1, Daniel Hebenstreit
Reviewed By
AnonymousHistory
Received: September 12, 2020Revision received: October 10, 2020
Accepted: October 16, 2020
Published: October 16, 2020
Copyright
© 2020 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Singh, AK; Zhang, J; Hebenstreit, D; Brogna, S (2020). Evidence of slightly increased Pol II pausing in UPF1-depleted Drosophila melanogaster cells. microPublication Biology. 10.17912/micropub.biology.000319.Download: RIS BibTeX