Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute (IDIBELL), 08908 L'Hospitalet de Llobregat, Barcelona, Catalonia, Spain
Center for Biomedical Research on Rare Diseases (CIBERER), ISCIII, Madrid, Spain.
Catalan Institution of Research and Advanced Studies (ICREA), Barcelona, Catalonia, Spain.
Faculty of Medicine, University of Vic-Central University of Catalonia (UVic-UCC), 08500 Vic, Spain
Institut de Neurociències, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain
Departament de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain
Abstract
X-linked Adrenoleukodystrophy (X-ALD) is a neurometabolic disorder caused by a defective peroxisomal ABCD1 transporter of very long-chain fatty acids (VLCFAs). We have characterized a nematode model of X-ALD with loss of the pmp-4 gene, the worm orthologue of ABCD1. These mutants recapitulated the key hallmarks of X-ALD and importantly mitochondria targeted antioxidant MitoQ prevented axonal degeneration and locomotor disability. In this study, we further demonstrated that the AWB chemosensory neuron of the pmp-4 mutant worm is defective, both in morphology and function. Interestingly, MitoQ could rescue both the phenotypes. Collectively, our results suggest that C. elegans’ chemosensation might provide a novel setting for exploring peroxisomal disease related disorders.
Description
X-linked adrenoleukodystrophy (X-ALD) is a rare neurometabolic disease characterized by inflammatory demyelination in the brain and axonal degeneration in the spinal cord. The disease is caused by mutations in the ATP binding cassette subfamily D member (ABCD1) gene, which encodes the peroxisomal transporter of very long chain fatty acids (VLCFAs) (Ferrer et al., 2010). Accumulation of VLCFAs, increased oxidative stress, and mitochondrial impairment are considered as the main etiological factor of X-ALD and other neurodegenerative disorders as reviewed in Kemp et al., 2016, Turk et al., 2020 and Guha et al., 2020 (a).
The C. elegans ortholog of human ABCD1 (and ABCD2) is PMP-4, encoded by the pmp-4 gene. We have established a novel in vivo model of X-ALD in C. elegans by using the mutant strain VC189 pmp-4(ok396). In this nematode model of X-ALD, PMP-4 protein was not expressed thus demonstrating that pmp-4(ok396) is a null allele (Coppa et al., 2020). In our previous manuscript we have demonstrated that PMP-4 absence causes VLCFA accumulation, severe neurodegeneration and production of high amount of mitochondrial reactive oxygen species (ROS) (Coppa et al., 2020). Interestingly, all these pathological phenotypes were rescued by treating worms with the mitochondrial targeted anti-oxidant drug, MitoQ (mitoquinone mesylate): which has the capability of reducing ROS generated by dysfunctional mitochondria (Coppa et al., 2020).
The role of peroxisomes in lipid metabolism nowadays is more focused towards ether phospholipid biosynthesis, fatty acid alpha-oxidation and fatty acid beta-oxidation (Wanders et al., 2017). In addition, peroxisomes also supply cholesterol to primary cilia, non-motile antenna-like protrusions that picks up signals required for embryonic development and adult tissue maintenance (Miyamoto et al., 2020).This links peroxisomal disorders with another group of human diseases called ciliopathies, characterized by cilia dysfunction (Waters et al., 2011).
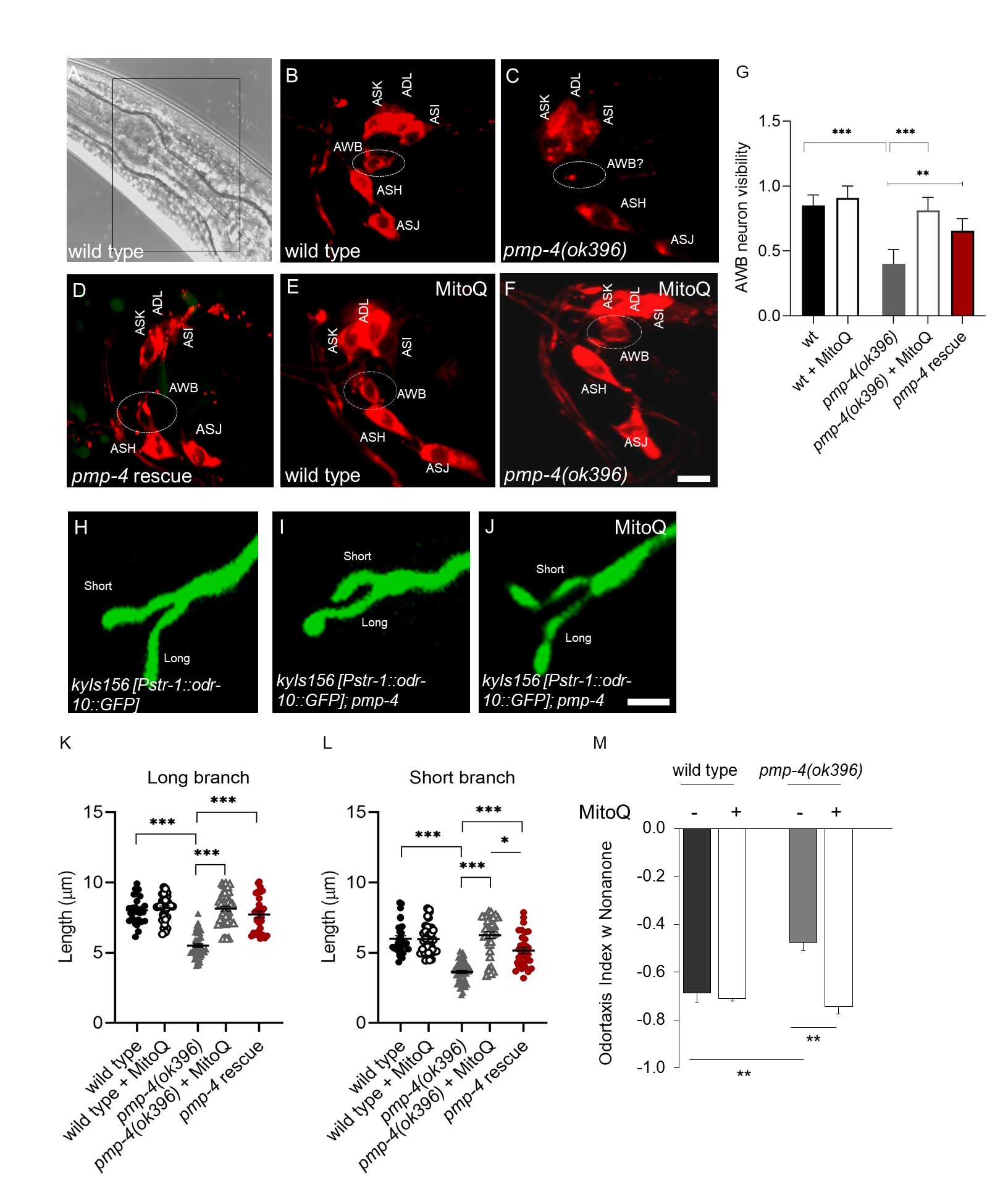
Human ciliopathies have been investigated in C. elegans by studying the functionality of the amphid organ, the chemosensory organ located in the head of the worm, composed of glia-like cells surrounding the chemosensory neurons (Stout et al., 2014; Shaham et al., 2015). Considering the possibility of establishing our model as a new platform for the investigation of ciliopathies and willing to go further along this new peroxisome-cilia association (Miyamoto et al., 2020), we deeply investigated the functionality of the amphid organ in the C. elegans model of X-ALD, constituted by the VC189 pmp-4(ok396) strain. In our recent study, we used two different approaches. First, the lipohilic dye 1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindocarbocyanine Perchlorate (‘DiI’; DiIC18(3), named DiI from now on, was used to visualize the dye filling behavior associated with chemosensory neurons functionality in the amphid organ (Schultz et al., 2012). Second, we used reporter strain named (CX3877 kyIs156 [Pstr-1::odr-10::GFP]) to visualize the structural morphology of the specific chemosensory neuron, Amphid Wing B (AWB) (Olivier Mason et al., 2013). kyIs expresses GFP-labeled ODR-10 in STR-1 (a Seven Transmembrane Receptor involved in maintaining cell to cell connection) expressing cells (Mukhopadhyay et al., 2008; Troemel et al., 1997). Results obtained with the DiI stain, showed that six pairs of neurons in the head amphid sensory organs fill with DiI in wild-type animals (Fig. 1A, B). However, we found that inactivation of PMP-4 caused failure of dye uptake, specifically in the AWB neuron, but had no effect on the adjacent DiI sensitive neurons (Fig 1C). Interestingly, this dye-filling defect of pmp-4 mutants was no longer detected by the rescue strain, containing PMP-4::GFP driven by the pmp-4 promoter and injected in pmp-4(ok396) animals; pmp-4 (ok396) [Ppmp-4::pmp-4(cDNA)::GFP] ( and named rescue strain from now on for clarity) (Fig 1D) thus confirming the specificity of this dye-filling phenotype. Our previous study demonstrated that mitochondrial ROS were responsible of X-ALD associated phenotypes (Coppa et al., 2020). Accordingly, we hypothesized and further investigated the in vivo ability of the mitochondria-targeted antioxidant, MitoQ, to protect against ROS-induced chemosensory neuronal damage in the pmp-4 mutant animals. Worms were treated separately with the MitoQ (at 5 µg/ml) and the DiI uptake by the AWB neuron of the anti-oxidant treated worms were analyzed. Astonishingly, AWB DiI staining was rescued in all MitoQ treated pmp-4 mutant animals (Fig 1 E, F, and G).
DiI dye-filling defect are frequently associated with defects in cilia or dendrite morphology (Olivier Mason et al., 2013; Ou et al., 2007) and mutants with defects in cilia or dendrite morphology frequently exhibit dye-filling defects in some or all dye-filling neurons (Olivier Mason et al., 2013). To determine whether the observed dye-filing phenotype in the pmp-4 mutant animals reflected defects in the neuronal morphology, we studied the AWB cilia structure by crossing the reporter strain kyIs156 [Pstr-1:: odr-10::GFP],with pmp-4(ok396) mutant animals (Fig 1H). Generally all AWB cilia in wild type animals possess the characteristic Y-shaped structure containing two branches of different lengths (Fig 1H), the distal ends of which exhibit an irregular morphology and exhibit some animal to animal variability in the lengths of each cilial branch, depending upon which temperature they are grown and in which conditions (presence or absence of food) (Mukhopadhyay et al., 2008).
However, pmp-4 mutant animals showed both cilia branches slightly shorter than in wild-type animals (Fig. 1K, L) thus suggesting an underlying correlation between failures of DiI uptake with the regulation of cilia length (Fig. 1I, 1K, L). Interestingly, like the dye-filing defect, the AWB cilia branch length defects were recovered in the pmp-4 rescue strain (Fig. 1K and L). Therefore PMP-4 is essential to maintain AWB cilia morphology. As we demonstrated in our previous manuscript how MitoQ rescued X-ALD associated phenotypes (Coppa et al., 2020)., here also we observed that the same concentration of MitoQ also rescued the AWB distal cilia morphology in pmp-4 mutant worms (Fig 1 J, K, L). Finally, the functional behavior associated with the AWB neuron was measured by odortaxis assay with nonanone, which generally C. elegans avoid (Troemel et al., 1997; Tanimoto et al., 2017). Odortaxis index in pmp-4 mutant worms was decreased in comparison to wild types, and MitoQ treated animals showed a recovery in this index, as well (Fig 1N). Thus, through mitochondrial antioxidant treatment, we have demonstrated that MitoQ suppresses ROS-induced chemosensation defects in the pmp-4 (ok396) worms.
Apart from the X-ALD specific phenotypes described in Coppa et al., 2020, in this study we have discovered that pmp-4 mutant worms are also defective in AWB chemosensory neuron cilia structural integrity which can lead inadequate function and defective staining. This phenotype can be regulated by mitochondrial ROS, since the mitochondria specific anti-oxidant MitoQ completely rescues all the defective phenotypes. Collectively, our results suggest that C. elegans chemosensation phenotype can be used as a platform for the investigation of human ciliopathies in which peroxisomes are involved, and provides another setting to explore peroxisomal-related disorders.
Methods
Request a detailed protocolC. elegans strains growth and maintenance
Nematodes were maintained at 200C on Nematode Growth Media (NGM) plates made with Bacto Agar (BD Biosciences). The plates were seeded with live E. coli OP50-1 bacterial strain (cultured overnight at 37oC at 220 rpm) and allowed to grow overnight, as previously described in (Brenner S, 1974). For experimental assays, after synchronization by standard procedure with sodium hypochlorite, 4th larval stage (L4) hermaphrodites (characterized by the appearance of a “Christmas tree vulva”) were selected and used for all the experiments.
Dye Filling Assay
DiI: 1,1´- Dioctadecyl-3,3,3′,3′- Tetramethylindo carbocyanine Perchlorate (Aldrich) staining was performed as previously described in (Schultz et al., 2012 and Tong et al., 2010). Briefly, a stock dye solution of 2 mg/ml DiI in dimethyl formamide was stored at -20ºC in a tube wrapped in foil to avoid oxidation. L4-staged well-fed worms from a plate were transferred into an eppendorf tube with 1 ml M9, into which 5 ul Dil solution from the stock was added. Tubes were shielded from light with aluminum foil and incubations were carried out at room temperature on a slow shaker for overnight. Next day, worms were mounted on a thick layer of half-dried agar (3%) pad on microscopic glass slides and subjected to confocal microscopy.
Confocal Microscopy
For ciliary morphology, animals were grown at the appropriate temperature were mounted on agarose pads set on microscopic slides and anaesthetized using 50 mM sodium azide in water (Sigma). Confocal images were acquired using a Leica spectral confocal microscope equipped with 63X objectives. Cilia length and morphology measurements were performed using ImageJ software (National Institutes of Health), after Z-stacking all the images, as described in Guha et al., 2020(b).
MitoQ Assay
For rescue experiments worms were fed with MitoQ (mitoquinone mesylate): 10-(4,5-dimethoxy-2-methyl-3,6-dioxocyclohexa-1,4-dien-1-yl)decyl)triphenylphosphonium methanesulfonate, as described in (Ng et al., 2014; Coppa et al., 2020). One milligram of MitoQ10 was dissolved in 1 ml of distilled water and 250 ul (1mg/ml) was added to autoclaved and cooled to 60ºC NG agar medium (total 50_ml). Synchronized L1 worms were placed on the MitoQ and normal NGM agar plates and grown for 48 hours till they reach the L4 stage. In parallel the same experiment was performed in plates without MitoQ as a control. Finally, worms were washed of the plates with M9 buffer and similar DiL or confocal experiment was performed as mentioned before.
Odortaxis assays
Avoidance assays for 2-nonanone was performed on square plates as described previously (Troemel et al., 1997). Plates were divided into six sectors labeled A-F. One microliter each of odorant (10% Nonanone) and 1 M NaN3 were added in two spots in sector A, and 1 μl each of control diluent (absolute ethanol) and 1 M NaN3 were added in two spots in sector F. An avoidance index (AI) or odortaxis index was calculated as [(number of animals in sectors A and B) − (number of animals in sectors E and F)]/ Total number of animals in all six sectors of the plate.
Reagents
| Genotype | Name | Source | Comment |
| Wild type (WT) | N2 | CGC | |
| pmp-4(ok396) IV | VC189 | CGC | |
| pmp-4(ok396) IV | EDC1 | This study | 10X outcrossed |
| kyIs156 [Pstr-1::odr-10::GFP] | CX3877 | CGC | GFP expression, specifically in the AWB neuron |
| kyIs156 [Pstr-1::odr-10::GFP]; pmp-4 | EDC61 | This study | |
| ibbEx42[Ppmp-4::pmp-4::GFP::(pmp-4)3’UTR; rol-6(su1006)] | EDC8 | This study | |
| pmp-4(ok396) IV; ibbEx42[Ppmp-4::pmp-4::GFP::(pmp-4)3’UTR; rol-6(su1006)] | EDC10 | This study | rescue strain used to confirm the specific role of PMP-4 in ciliary abnormalities |
All the strains can be ordered on request from Prof. Dalfo’s lab which is situated in the Institut de Neurociències, Autonomous University of Barcelona, Bellaterra Campus, Catalonia, Spain.
GFP fluorescence was used to guide selection of progenies, and PCR genotyping was used to confirm homozygosity with primers specific to the ok396 deletion, including: 5´-TCGGTAATCCCTTGTCTTCTC-3´ (forward), 5´- CGGAGGTCATCAGGTTTGTT-3´ (reverse) and 5´- ACTCCGAAGCCGATGAAATT-3´ (deletion).
Acknowledgments
We thank Dr. Michael Murphy for sending us the MitoQ reagent. C. elegans strains used in this work was provided by the Caenorhabditis Genetics Center (CGC), funded by the NIH Office of Research Infra- structure Programs (P40OD010440). We thank the members of Dr. Pujol’s lab, members of the Spanish worm meeting group for their valuable suggestions and helpful discussions.
References
Funding
SG - FI Predoctoral Fellowship, AGAUR, Catalunya, Spain. ED - The Spanish Ministry of Science and Competitive grants (PC0009/003 and PI1100968)
Reviewed By
AnonymousHistory
Received: October 29, 2020Revision received: December 22, 2020
Accepted: January 14, 2021
Published: January 14, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Guha, S; Pujol, A; Dalfo, E (2021). Anti-oxidant MitoQ rescue of AWB chemosensory neuron impairment in a C. elegans model of X-linked Adrenoleukodystrophy. microPublication Biology. 10.17912/micropub.biology.000346.Download: RIS BibTeX