Abstract
To facilitate genetic mapping of developmental mutants of Physcomitrium patens, we produced a genetic marker that combines recessive auxotrophy with dominant positive selection. We first identified the gene affected by the pabB4 auxotrophic mutation and then replaced it with a cassette that confers antibiotic resistance. This strain may be used to produce bi-parental somatic hybrids with nearly any other strain.
Description
The haploid-dominant life cycle of mosses poses a challenge to genetically mapping infertile mutants. Recently, a method to circumvent these difficulties was developed that uses protoplast fusion of mutant and wild-type protoplasts to produce fertile somatic hybrids that produce segregating sporelings upon selfing (Moody et al. 2018). All known methods for producing somatic hybrids require the parent strains to either have complementing auxotrophic mutations or distinct antibiotic resistances (Grimsley et al. 1977; Cove et al. 2009b). Here we describe a strain that could be used as a universal fusion partner for mutants that contain neither type of marker, one that combines recessive auxotrophy with dominant antibiotic resistance.
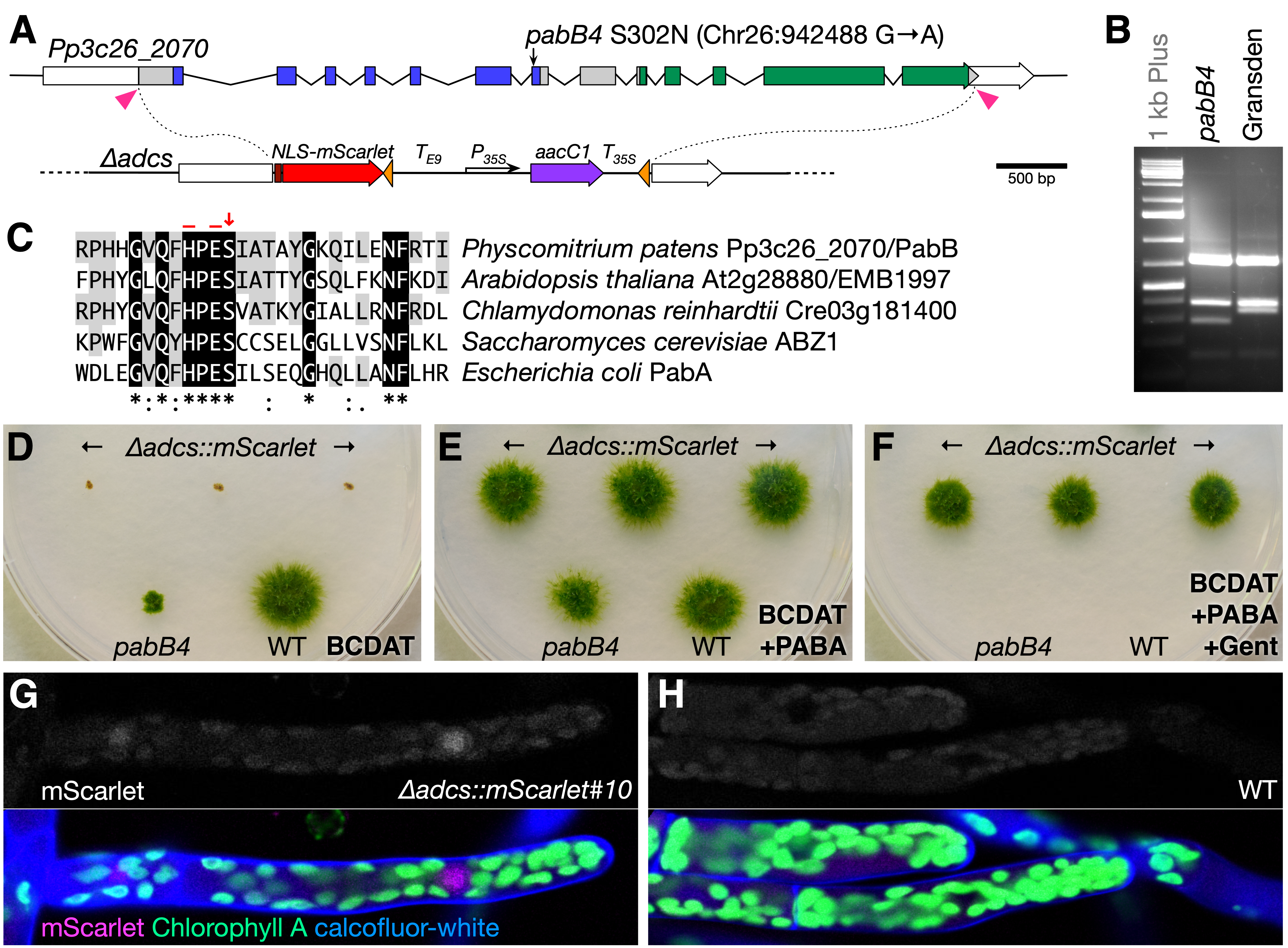
Among the most commonly used auxotrophic mutants of the moss Physcomitrium patens (Hedw.) Mitt. (previously Physcomitrella patens) are those that require p-Aminobenzoate (PABA) for growth and which fall into two complementation groups, pabA and pabB (Ashton and Cove 1977; Grimsley et al. 1977; Ashton et al. 1979). PABA, along with pterin and glutamate moieties, is essential in the production of folates (Vitamin B9) which, in turn, are essential cofactors for one-carbon transfer reactions in the synthesis of various compounds such as methionine, purines, and thymidylates (reviewed in Hanson and Roje 2001). PABA is synthesized from chorismate and glutamine in three steps. The first two steps, glutamine hydrolysis and adding the resulting amino group to chorismate, are catalyzed by the Aminodeoxychorismate Synthase (ADCS) enzyme comprised of a single bifunctional protein in most plants and fungi and by separate glutaminase and synthase subunits in most bacteria (Basset et al. 2004a). (Note that the P. patens pabA and pabB complementation groups were named independently from the bacterial PabA and PabB enzyme subunits.) The 4-amino-4-deoxychorismate (ADC) product is converted to PABA by the ADC Lyase enzyme (Basset et al. 2004b).
In P. patens, ADCS is encoded by a single gene, Pp3c26_2070, whereas three genes (Pp3c2_23040, Pp3c4_31240, and Pp3c7_15160) appear to encode ADC lyase enzymes. We sequenced the Pp3c26_2070 locus from the pabB4 mutant and found a single mutation in the seventh exon that results in an asparagine substitution at a highly conserved serine residue (S302N) in the glutaminase domain immediately adjacent to the His-299 and Glu-301 active site residues and also creates an SspI restriction site (Fig. 1A–C). The corresponding serine in the bacterial glutaminase subunit was shown to form two critical hydrogen-bonds that link an Asp residue of the synthase subunit to a Thr residue of the glutaminase subunit; interactions between these three residues mediate the allosteric stimulation of glutaminase activity by chorismite binding to the synthase subunit (Semmelmann et al. 2019). Interestingly, the Asp residue is conserved in plant ADCS enzymes, but the Thr is not. It is not currently known whether chorismate binding also stimulates glutaminase activity in plant ADCS enzymes.
To confirm that Pp3c26_2070 is required for PABA synthesis, we replaced its coding region in a wild-type strain’s genome with a cassette conferring resistance to gentamicin and a nuclear-localized mScarlet fluorescent-protein gene using CRISPR/Cas9-facilitated targeted gene replacement (Fig. 1A). Sixty-seven of seventy-two stable transformants assayed grew only on media supplemented with PABA, and three lines with clean gene replacements were selected based on PCR genotyping (∆adcs::mScarlet; Fig. 1D–1F). By comparison, the non-null pabB4 mutant grew very slowly without added PABA (Fig. 1D and 1E). Weak fluorescent signal from mScarlet could be detected above background chloroplast-derived autofluorescence (Fig. 1G versus 1H).
The ∆adcs::mScarlet mutant is potentially a useful genetic tool. It was designed to extend the mutant mapping system developed by Moody et al. (2018) to mutants whose background lack an antibiotic resistance that could be selected. After fusion with ∆adcs::mScarlet, only bi-parental hybrids would be able to grow on minimal media supplemented with gentamicin—the mutant’s genome would provide a functional ADCS gene and ∆adcs::mScarlet’s genome would confer gentamicin-resistance. Such a universal hybridization partner might also allow production of allopolyploid lines through fusion with protoplasts from other moss species, most of which lack established transformation protocols. Auxotrophic mutants may also prove essential in the development of stably maintained shuttle vectors in moss. Unlike other plants, P. patens can maintain plasmid DNA extrachromosomally as long as selection is applied (Ashton et al. 2000; Murén et al. 2009). Vectors that complement auxotrophic mutations may be superior to those that confer antibiotic resistance because selection can be maintained even in cells not in direct contact with the substrate (Ulfstedt et al. 2017).
Methods
Request a detailed protocolMoss propagation and transformation were carried out as described previously (Cove et al. 2009a; Cove et al. 2009c). The Pp3c26_2070 gene was amplified and sequenced in four segments using the indicated primers (Primer Table) from the pabB4 mutant (kindly provided by Neil Ashton, University of Regina). To confirm that the G-to-A mutation in the seventh exon was unique to the pabB4 genome, the region including the mutation was amplified from pabB4 (Gransden background) and the ‘Gransden 2004’ wild type and digested with SspI. The SspI site created by the G-to-A mutation was only present in the pabB4 product.
The ∆adcs::mScarlet construct, pMP1907, was created by ligating in SpeI–SwaI and SphI–SalI fragments with the downstream and upstream homology arms, respectively, into the pMP1119 vector. pMP1119 was created from pBNRF (Thelander et al. 2007) by 1) digestion with BglII and NotI followed by polishing with T4 DNA polymerase and re-ligation, 2) removal of the 35S:nptII transgene by EcoRI digestion and re-ligation, and 3) insertion of the 35S:aacC1 transgene as a KpnI–SacI fragment after amplifying from pYL-TAP-Nt (Rubio et al. 2005). The Pisum rbcS-E9 terminator sequence was inserted as a KpnI fragment upstream from the 35S:aacC1 transgene. NLS-mScarlet (Bindels et al. 2017) was amplified and subcloned into pCR Blunt (ThermoFisher) then inserted as an EcoRI fragment into the MfeI site. The plasmid to express SpCas9 and guide RNAs designed to target near the start- and stop codons (pMP1957) was generated using oligos and pMK-Cas9-gate according to published protocols (Mallett et al. 2019). Fifteen µg of both pMP1907 and pMP1957 plasmids were transformed into the Reute 2016 strain (Hiss et al. 2017). After regeneration on PRMB medium containing 3 µM PABA, four-day-old transformed protoplasts were selected on BCDAT medium containing 3 µM PABA, 100 mg/l gentamicin, and 20 mg/l G418 for one week. Transformants were picked to BCDAT+PABA medium, then 10 days later a small clump of each was transferred to BCDAT+PABA+gentamicin medium to identify stable transformants. We later discovered that expression of aacC1 confers resistance to both gentamicin and G418 (but not to 100 mg/l kanamycin), however the high rate of stable integration likely reflects the high rate of co-transformation despite no selection for pMP1957 uptake. The presence of proper 5′ and 3′ integration products and the absence of SpCas9 and ADCS genes were confirmed by PCR (Primer Table).
mScarlet, calcofluor-white, and chlorophyll fluorescent signals were imaged using a Zeiss LSM 880 microscope using 561, 405, and 633 nm excitation and 580–605, 410–501, and 647–721 nm detection windows, respectively.
Reagents
| Primer Table | ||
| Name | Sequence (5′ to 3′) | Purpose |
| P35S-KpnF | atcggtaccAACATGGTGGAGCACGAC | Subcloning 35S:aacC1 |
| T35S-SacR | tcggagctcCTGGATTTTGGTTTTAGGAATTAGA | Subcloning 35S:aacC1 |
| SV40FP-XbaF | tctagaATGGCTCCAAAGAAGAAGAGAAAGGTCGCTGTGAGCAAGGGCGAGGA | Subcloning NLS-mScarlet |
| mCherry-xbaR | tactctagaTTACTTGTACAGCTCGTCCATGC | Subcloning NLS-mScarlet |
| Te9-KpnF | TCCggtaccGTTCGAGTATTATGGCATTGGG | Subcloning rbcS-E9 |
| Te9-KpnR | GTTgGtACcATTGGCAAGTCATAAAATGCATT | Subcloning rbcS-E9 |
| ADCS5-SphF | CAAgcATGCTTTTTTTCAAAGCAAATTTG | Subcloning 5′ targetting arm |
| ADCS5-SalR | TGCgtCGACCTCAAGCTCCATTTTCAGACC | Subcloning 5′ targetting arm |
| ADCS-3SpeF | GAAacTAGTGTGGCTTTACCTTAGTCTCCTC | Subcloning 3′ targetting arm |
| ADCS-3R | AACACCTTCACTTATATGCCTCCA | Subcloning 3′ targetting arm |
| ADCS-5-crF | ccatGCACCTGGAGATGACTCAGA | CRISPR protospacer |
| ADCS-5-crR | aaacTCTGAGTCATCTCCAGGTGC | CRISPR protospacer |
| ADCS-3-crF | ccatACACCACCTCCAGCAGTCAA | CRISPR protospacer |
| ADCS-3-crR | aaacTTGACTGCTGGAGGTGGTGT | CRISPR protospacer |
| ADCS-5uF | GGTCTGAAAATGGAGCTTGAGGT | Sequencing |
| ADCS-4iR | AGGAGAAGAAGGAGCAAAGCAGA | Sequencing |
| ADCS-4iF | CGGTCGTTTAAGGTATAATTTCTCCA | Sequencing, pabB4 genotyping |
| ADCS-8eR | ATCAGAACATGGCTTGAATCGTC | Sequencing, pabB4 genotyping |
| ADCS-8eF | GATCTTACGAAGTGCCTGCATGA | Sequencing |
| ADCS-12eR | TAGAGAGCCATTCGACTTGGAAAC | Sequencing |
| ADCS-12eF | ATTCGTTTAATCACGGCCAGAAC | Sequencing |
| ADCS-3uR | ATCCCCTGATGGAACTACGTGAA | Sequencing |
| Ubi-tataF2 | CGATGCTCACCCTGTTGTTTGG | ∆adcs genotyping (Cas9) |
| Cas9-R | TTGATCATGGAGGCGGAGAGTG | ∆adcs genotyping (Cas9) |
| ADCS-genoF | GGATAGAGCCCCACAAAGCCA | ∆adcs genotyping (5′) |
| NLS-genoR | ACCTTTCTCTTCTTCTTTGGAGCCA | ∆adcs genotyping (5′) |
| Tcamv-genoF | CCTATAGGGTTTCGCTCATGTGTTG | ∆adcs genotyping (3′) |
| ADCS-genoR | CCAATAAGTCCTACCAAATAAACGCCT | ∆adcs genotyping (5′) |
| ADCS-e6F | ATGCTCCTGGGGTTGATTGCT | ∆adcs genotyping (WT) |
| ADCS-e11R | CACAAAAGTGGAAGGGCAGGC | ∆adcs genotyping (WT) |
Acknowledgments
We thank Neil Ashton for providing the pabB4 mutant and for discussions about PABA-requiring mutants.
References
Funding
Work in the Estelle lab was supported by NIH (GM43644).
Reviewed By
Luis VidaliHistory
Received: December 23, 2020Accepted: January 23, 1970
Published: January 26, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Prigge, MJ; Wang, Y; Estelle, M (2021). Mutations in the Physcomitrium patens gene encoding Aminodeoxychorismate Synthase confer auxotrophic phenotypes. microPublication Biology. 10.17912/micropub.biology.000364.Download: RIS BibTeX