Abstract
Delineated as the first cellular organelle in 1675 by Antonie van Leeuwenhoek, cilia did not receive much attention until the 2000s, when it became apparent that cilia played a key role in the development of embryos, a variety of signaling pathways. Therefore, collective efforts by many scientists have led to the identification of many novel ciliopathy and cilia genes, while we are still far from disclosing the complete components of cilia. Here we used the ciliated sensory neurons in C. elegans as a model system that revealed the voltage-gated K+ channel EGL-36 (a member of the Shaw subfamily) as a new component associated with cilia. The confocal microscopy examination of fluorescence tagged EGL-36 together with ciliary (IFT-140) or transition zone (MKS-6) markers reveal that EGL-36 is only expressed in subsets of the ciliated sensory neurons, where it partially overlaps with the basal body signals and predominantly localizes to the periciliary membrane compartment. This expression pattern along with studies of egl-36 gain-of-function variants indicates that egl-36 is not essential for ciliogenesis in C. elegans. Our data identify the voltage-gated K+ channel EGL-36 as a new cilia-associated protein, and future studies should reveal the functional significance of EGL-36 in cilia biogenesis.
Description
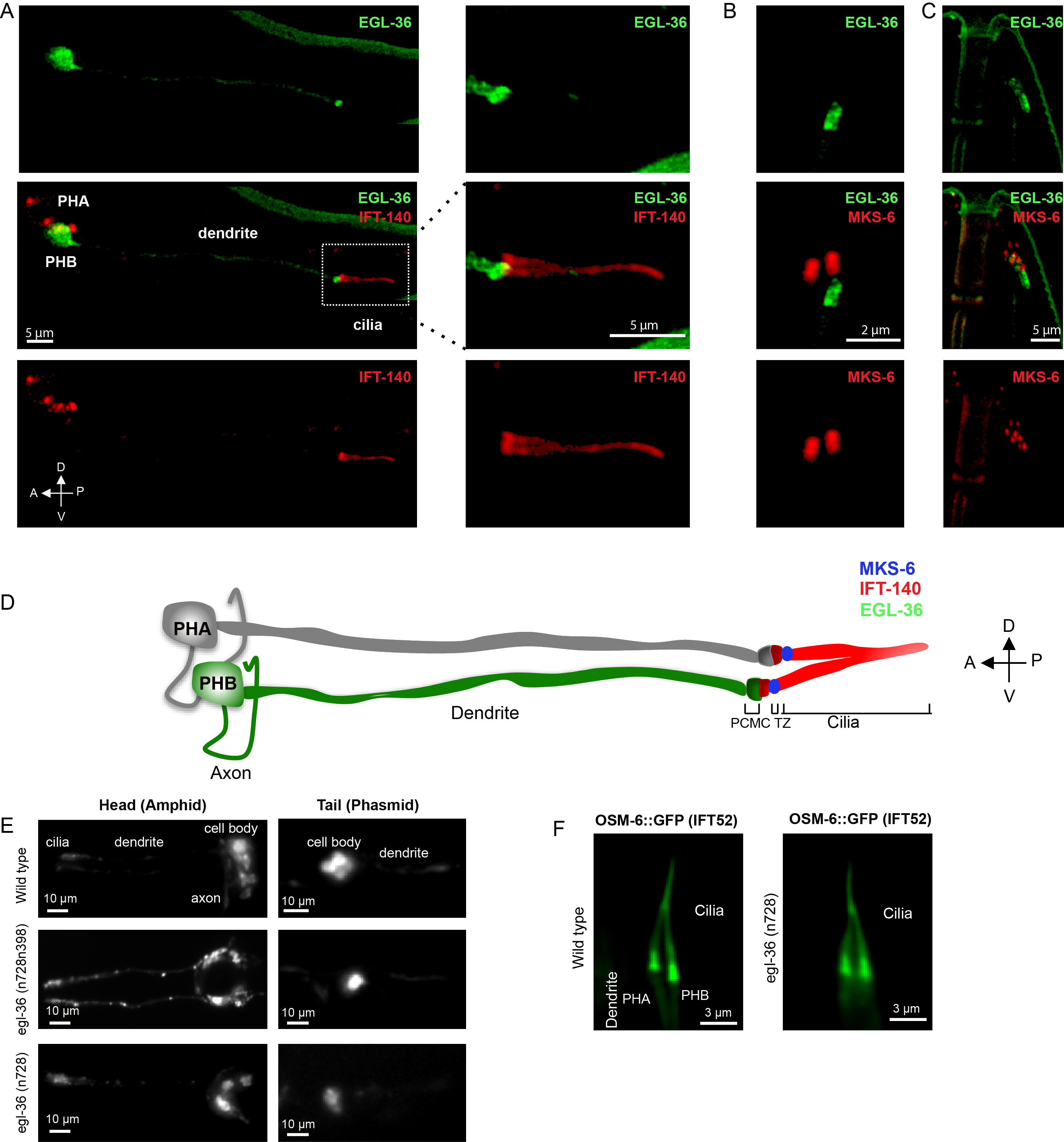
C. elegans EGL-36 displays extensive similarity to human KCNC proteins, the conserved potassium channel subfamily KV3 (the Shaw subfamily). This potassium channel subfamily contains four genes KCNC1, KCNC2, KCNC3, and KCNC4 encoding KV3.1 (Shaker), KV3.2 (Shab), KV3.3 (Shaw), and KV3.4 (Shal), respectively. Previous studies have demonstrated that C. elegans EGL-36 is expressed in the egg-laying muscles, ventral cord neurons, and the subset of ciliated sensory neurons including ADEL, ADER, PHB through its subcellular localization in the ciliated sensory neurons remains unknown (Elkes et al. 1997; Johnstone et al. 1997). To observe the subcellular localization of EGL-36 in the ciliated sensory neurons in C. elegans, we tagged green fluorescent protein (GFP) at the C-termini of EGL-36 with a cilia-specific promoter. We observed overexpressed EGL-36::GFP in the subsets of sensory neurons in the head and tail including PHB sensory neurons in the tail but not in the PHA sensory neuron, which is in line with previous findings (Elkes et al. 1997; Johnstone et al. 1997) (Figure 1A). Confocal laser scanning microscopy shows that EGL-36::GFP is predominantly located at the base of the cilium. The EGL-36::GFP signal can be found in the neuronal cell body, with weak dendrite staining, where they may function in the dendritic excitability (Figure 1A).
Cilia have several sub-compartments including the basal body, the transition zone, the middle segment, and the ciliary tip. We further examined the exact localization of GFP-tagged EGL-36 relative to these compartments. To this end, we used the transition zone marker MKS-6/CC2D2A and the cilia marker IFT-140 (intraflagellar transport 140), and co-expressed EGL-36::GFP with either of these markers (Qin et al.. 2001; Williams et al.. 2011; Prevo et al. 2015 Mijalkovic et al. 2018). Our microscopy analysis revealed that some of EGL-36::GFP signal was, however, found to overlay with the proximal zone of IFT signal (marked with IFT-140::mCherry), while the majority of EGL-36:: GFP signal was predominantly localized in a compartment, proximal to the ciliary base (Figure 1A). This strong EGL-36 staining likely corresponds to the periciliary membrane compartment (PCMC) (Kaplan et al. 2012). There is no overlay of EGL-36::GFP signal with MKS-6::mCherry (Figure 1B and C).
We next went on characterizing potential ciliary functions of EGL-36 in C. elegans, we therefore obtained two mutants for egl-36 including egl-36(n728) and egl-36(n728n398). egl-36(n728) mutant contains a missense variant, conversion of glutamic acid at the 142nd position of EGL-36 to lysine (p.E142K). EGL-36(p.E142K) represents a gain of function variant of egl-36, which displays defects in egg-laying in C. elegans. On the other hand, proline to serine conversion (p.P439S) in egl-36 suppresses the egg-laying defect of p.E142K variant (Elkes et. al., 1997; Johnstone et. al., 1997). egl-36(n728n398)] mutant contains both p.E142K and p.P439S variants. First, we performed the DiI-filling assay, which has been used to test ciliary structural defects in C. elegans. The lipophilic fluorescent dye is not taken up by C. elegans with abnormal cilia structures, while wild types always stain their ciliated sensory neurons with this fluorescent dye (Herman and Hedgecock 1990). The dye analysis coupled with microscopy imaging revealed that similar to wild type, both egl-36 mutants fill up their ciliated sensory neurons with the lipophilic fluorescent dye, suggesting that egl-36 mutants have no gross cilia abnormalities in C. elegans (Figure 1E). We next examined the cilia morphologies of phasmid (tail) sensory neurons in egl-36(n728) mutant. We generated transgenic strains expressing OSM-6/IFT52::GFP in egl-36(n728) mutants, and our microscopy analysis revealed that PHB (tail) displayed no abnormality in cilia morphology in egl-36(n728) mutants (Figure 1F).
Methods
Request a detailed protocolC. elegans strains and maintenance
All strains used were grown in the nematode growth medium (NGM) at 20°C as previously described (Brenner, 1974).
Generation of transgenic strains with microinjection
The transgenic strain OIK904, N2;turEx12[Parl-13::EGL-36::GFP::unc-54 3`UTR+pRF4] was generated with microinjection of the Parl-13::EGL-36 (cDNA)::GFP::unc-54 3` UTR construct (25 ng/μl) along with the rol-6 co-injection marker (50 ng/μl) by our laboratory.
Dye-filling assays
Dye filling assay previously described was used (Cevik et al.. 2010; Herman and Hedgecock 1990).
Fluorescent and Confocal microscopy
Fluorescent images for the dye assay were obtained by a fully automated upright microscope system (Leica DM6 B ) with a Plan ApoChromat 100x/1.40 NA and an electron-multiplying charge-coupled device camera (Andor iXon Ultra 897 EMCCD camera and iQ3.6.2 Andor software) that is attached to the microscope while confocal images were acquired with a laser-scanning confocal inverted microscope (Zeiss LSM 900 with Airyscan 2 and ZEN 3 Blue edition software) with a Plan ApoChromat 63x/1.40 NA objective.
Reagents
Strains and plasmid are available upon request to sebiha.cevik(at)agu.edu.tr
| Strain | Genotype | Available from |
| N2 | Caenorhabditis elegans | CGC |
| MT1540 | egl-36(n728) X. | CGC |
| KP100 | egl-36(n728n398) X. | CGC |
| SP2101 | osm-6(p811);mnIs17[osm-6::gfp; unc-36(+) | CGC |
| OIK912 | egl-36 (n728)X.; mnIs17[osm-6::gfp; unc-36(+). | This study |
| OIK904 | turEx12[Parl-13::EGL-36::GFP::unc-54 3`UTR+pRF4] | This study |
| EJP81 | vuaSi24 [pBP43; Pche-11::che-11::mCherry; cb-unc-119(+)]II; unc-119(ed3) III; che-11(tm3433)V. (This is referred IFT-140 throughout to the text) | Peterman Lab |
| DAM954 | vuaSi21[pBP39; Pmks-6::mks-6::mCherry; cb-unc-119(+)]II.;mks-6(gk674) I (This is referred MKS-6 throughout to the text) | Dammermann Lab |
| OIK952 | N2;turEx12[Parl-13::EGL-36::GFP::unc-54 3`UTR+pRF4];vuaSi21[pBP39; Pmks-6::mks-6::mCherry; cb-unc-119(+)]II. | This study |
| OIK953 | N2;turEx12[Parl-13::EGL-36::GFP::unc-54 3`UTR+pRF4];vuaSi24 [pBP43; Pche-11::che-11::mCherry; cb-unc-119(+)]II; unc-119(ed3) III; che-11(tm3433)V. | This study |
| Plasmids | Genotype | Description |
| OK133 | Parl-13::EGL-36 (cDNA) ::GFP::unc-54 3`UTR | Inserted Parl-13 (300 bp)- egl-36 (cDNA) with SphI and Age I restriction enzymes by Sunybiotech |
| pRF4 | rol-6(su1006) | (Mello C et al. 1991) |
Acknowledgments
We thank Ferhan Yenisert for her technical help with microinjections. We thank Erwin J.G. Peterman and Alexander Dammermann for providing reagents. Some of C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We acknowledge that this work was partly funded by Scientific and Technological Research Council of Turkey (TUBITAK) Grant (Project number: 118Z471) to O.I.K
References
Funding
Scientific and Technological Research Council of Turkey (TUBITAK) Grant (Project number: 118Z471)
Reviewed By
AnonymousHistory
Received: January 29, 2021Revision received: February 19, 2021
Accepted: February 23, 2021
Published: March 5, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Cevik, S; Kaplan, OI (2021). Subcellular localization of the voltage-gated K+ channel EGL-36 , a member of the KV3 subfamily, in the ciliated sensory neurons in C. elegans. microPublication Biology. 10.17912/micropub.biology.000367.Download: RIS BibTeX