Abstract
Unpaired1 (Upd1) is a ligand of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway in Drosophila. In this study, using the CRISPR/Cas9 technique, we generate a transgenic fly strain in which a hemagglutinin (HA) epitope tag sequence is inserted into the endogenous locus of the upd1 gene. Anti-HA antibody staining confirms that the distribution of the epitope-tagged Upd1::HA in various tissues is consistent with upd1 expression patterns revealed by previous studies. This transgenic fly strain will be useful in studying the expression, localization, and association partners of Upd1, and thus will contribute to understanding how activation of the JAK/STAT pathway is regulated.
Description
The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway is involved in various biological processes, including cell proliferation, differentiation, and apoptosis (Morris et al. 2018). The core components of the JAK/STAT pathway are evolutionarily conserved between humans and flies (Arbouzova and Zeidler 2006). While more than 50 cytokines activate the JAK/STAT pathway in humans, there are only three JAK/STAT ligands in Drosophila (Morris et al. 2018). Unpaired 1 (Upd1), one of the three ligands of the Drosophila JAK/STAT pathway, is involved in many developmental processes, including growth and patterning of the wing disc (Ayala-Camargo et al. 2013; Recasens-Alvarez et al. 2017), stem cell maintenance in the testis (Cuevas and Matunis 2011), and specification of follicular epithelial cell fates in the ovary (McGregor et al. 2002).
Since Upd1 interacts with the extracellular matrix (Harrison et al. 1998; Hayashi et al. 2012), it is essential to understand its distribution and association partners at a protein level using antibodies. Polyclonal antibodies against Upd1 have been generated previously (Harrison et al. 1998; Zhang et al. 2013; Beshel et al. 2017). However, since polyclonal antibodies are typically purified from the serum of the immunized animal, their supply is limited and exhaustible.
Epitope tagging of endogenous protein is an alternative effective means for studying the expression, localization, and interactive association of a protein under normal and pathological conditions. Once an epitope tag sequence is successfully inserted into an endogenous gene locus, the tagged protein can be readily detected or purified using a commercially-available well-characterized monoclonal antibody for the epitope. In this study, we generated a knock-in strain in which an epitope tag is inserted into the endogenous locus of upd1 using CRISPR/Cas9-mediated homology-directed repair.
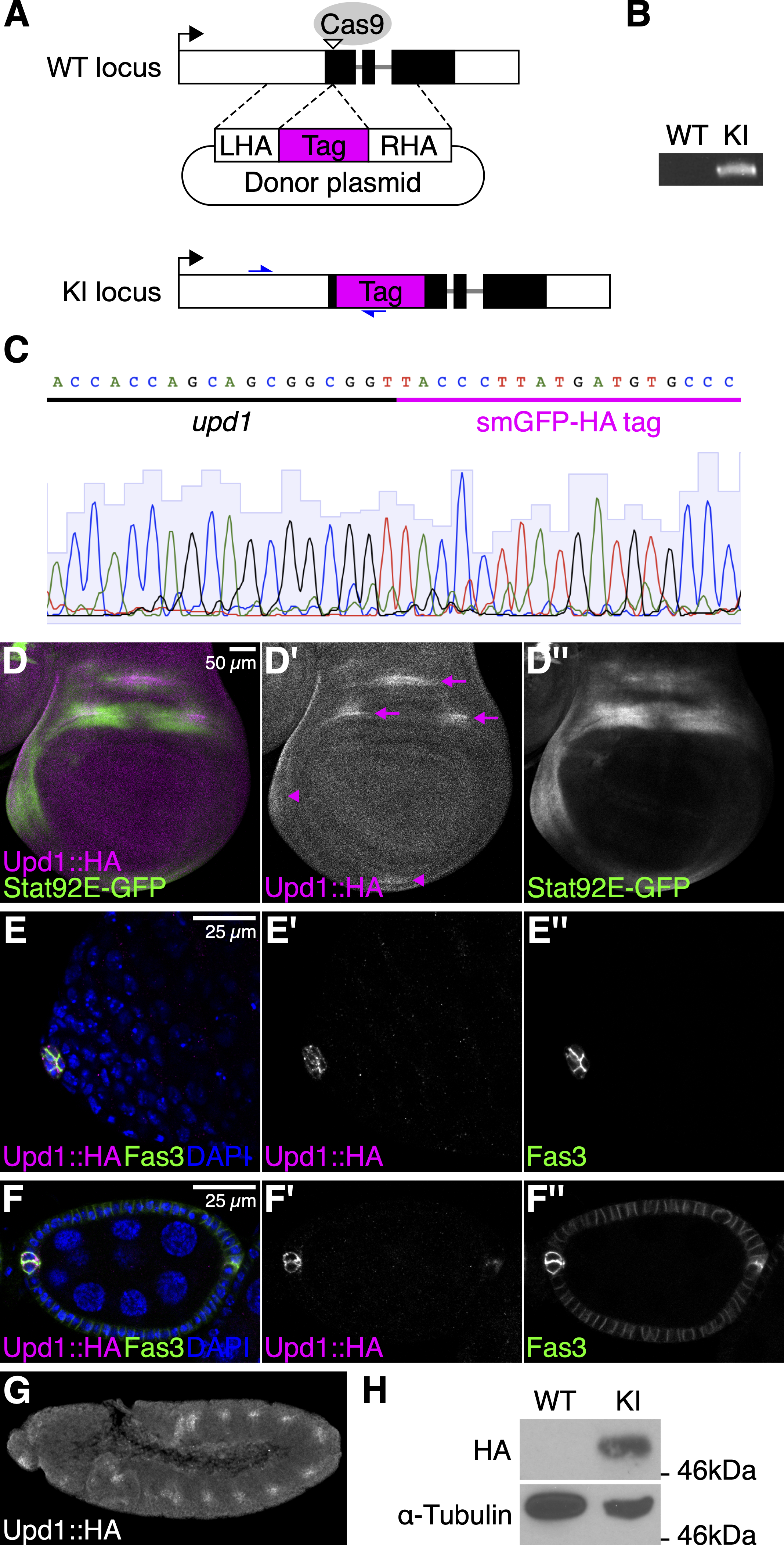
Upd1 has multiple potential furin cleavage sites (Arg133, Lys332, Arg402, and Arg405), which are predicted by the ProP v.1.0 server with relatively higher scores (≥0.45; Duckert et al. 2004). We inserted the non-fluorescent GFP-like protein containing 10 copies of influenza hemagglutinin (HA) epitope tag (smGFP-HA; Viswanathan et al. 2015) after Gly35 of Upd1, downstream of the N-terminal signal sequence (Fig. 1A). Flies with tag integration were screened by PCR using a tag-specific and genomic primer pair (Fig. 1B). Sanger sequencing was performed for validating precise integration (Fig. 1C). Here, we refer to this transgenic strain as upd1KI.HA and its protein product as Upd1::HA. The upd1KI.HA flies are viable and fertile.
We examined the expression patterns of the HA-tagged Upd1 from the upd1KI.HA flies in the wing disc, ovary, testis, and embryo using an anti-HA antibody. In the wing disc, we found that Upd1::HA was expressed within three regions of the presumptive dorsal wing hinge as well as in the anterior and ventral body wall (Fig. 1D). This observation is consistent with a previously-reported result determined by in situ hybridization (Johnstone et al. 2013). We also confirmed that using a Stat92E-GFP reporter (Bach et al. 2007), the JAK/STAT pathway was activated in surrounding regions of the Upd1::HA-expressing cells (Fig. 1D). Similarly, the expression of Upd1::HA was detected in a group of quiescent somatic niche cells (called hub cells) in the adult testis (Fig. 1E) and two pairs of polar cells at each end of the egg chamber in the adult ovary (Fig. 1F), as previously reported (Silver and Montell 2001; Tulina and Matunis 2001; Beccari et al. 2002). In the stage 11-12 embryo, we confirmed that Upd1::HA is expressed in the tracheal pits, consistent with the previously-reported pattern of upd1 mRNA (Fig. 1G; Harrison et al. 1998). Western blotting of embryonic extracts found that Upd1::HA was detected as a band at around 50kDa (Fig. 1H). This band size is consistent with the cleavage at Arg133, although biochemical analysis is required to determine the precise cleavage site. Taken together, these results demonstrate that the expression of Upd1 is faithfully detected using this upd1KI.HA fly strain. This strain will also be useful in studying the localization and association partners of Upd1, and thus will contribute to future research on the JAK/STAT pathway.
Methods
Request a detailed protocolDrosophila strain construction
The upd1KI.HA fly strain was generated using CRISPR/Cas9-mediated gene editing as previously described (Gratz et al. 2014; Takemura et al. 2021). Briefly, to construct a single guide RNA (sgRNA) plasmid, 5′-CTTCGAACCGTTAGACCGCCGCTGC-3′ and 5′-AAACGCAGCGGCGGTCTAACGGTTC-3′ were annealed and ligated in the BbsI-digested pU6-BbsI-chiRNA plasmid (a gift from Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger; Addgene #45946). We constructed a donor plasmid containing 757-bp and 975-bp arms homologous to sequences in the upd1 gene, flanking smGFP-HA and a Gly-Gly-Ser linker. A mixture of 50 ng/µl of the sgRNA plasmind and 250 ng/µl of the donor plasmid was injected into the vasa-Cas9 embryos by BestGene Inc. The upd1KI.HA allele was screened by PCR and verified by Sanger sequencing.
Immunohistochemistry
Dissection and antibody staining were performed as previously described (Takada et al. 2015; Takemura et al. 2020). The following primary antibodies were used: rabbit monoclonal anti-HA C29F4 (1:1000, Cell Signaling Technology, #3724) and mouse monoclonal anti-Fas3 7G10 (1:50, Developmental Studies Hybridoma Bank [DSHB], deposited by Corey Goodman). Alexa548- and Alexa633-conjugated secondary antibodies (Thermo Fisher Scientific) were used at a dilution of 1:500. Nuclei were stained with 1 µg/ml DAPI (Thermo Fisher Scientific, #62248). Images were acquired on an LSM710 confocal microscope (Carl Zeiss).
Western blotting
Embryonic protein extraction and western blotting were performed as previously described (Kleinschmit et al. 2013; Takada et al. 2015). The following antibodies were used: rabbit monoclonal anti-HA C29F4 (1:1000, Cell Signaling Technology, #3724), mouse monoclonal anti-α-Tubulin DM1A (1:1000, Sigma-Aldrich), anti-rabbit HRP-linked IgG (1:10000, Cell Signaling Technology) and anti-mouse HRP-linked IgG (1:10000, Cell Signaling Technology).
Reagents
The following fly stocks were used in this study:
| Strain | Genotype | Available from |
| vasa-Cas9 | y[1] M{GFP[E.3xP3]=vas-Cas9.RFP-}ZH-2A w[1118] | Bloomington Drosophila Stock Center (BDSC) #55821 |
| Stat92E-GFP | w[1118]; P{w[+mC]=10XStat92E-GFP}1 | BDSC #26197 |
| upd1KI.HA | upd1[KI; smGFP-HA] | This study |
Acknowledgments
We thank Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger for plasmids, and the BDSC (NIH P40OD018537) for fly stocks, and the DSHB for an antibody.
References
Funding
This work was supported by the National Institute of Health (NIH) grant R35 GM131688 (to H.N.).
Reviewed By
AnonymousHistory
Received: April 6, 2021Revision received: August 25, 2021
Accepted: August 26, 2021
Published: August 31, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Takemura, M; Lu, YS; Nakato, E; Nakato, H (2021). Endogenous epitope tagging of a JAK/STAT ligand Unpaired1 in Drosophila. microPublication Biology. 10.17912/micropub.biology.000387.Download: RIS BibTeX