Prodigy Biotech, Jupiter, FL 33458
ScienCell Research Laboratories, Inc., 1610 Faraday Avenue, Carlsbad, CA 92008
Abstract
In rodents, all three paralogs of the Attractin (Atrn) transmembrane protein family exhibit strong phenotypic overlap and are implicated in the regulation of the same G-protein coupled receptors (GPCR) as E3-ligase Mahogunin ring finger 1 (Mgrn1). Recently it was shown that the highly conserved intracellular MASRPF motif in mammal Multiple epidermal growth factor-like domain 8 protein is required for binding of Mgrn1 to mediate ubiquitination of GPCR Smoothened in vitro. Here, we show that the MASRPF motif of Drosophila Distracted, the ortholog of ATRN and Attractin-like 1, is required for association with Drosophila Mgrn1 (dMgrn1) in vivo.
Description
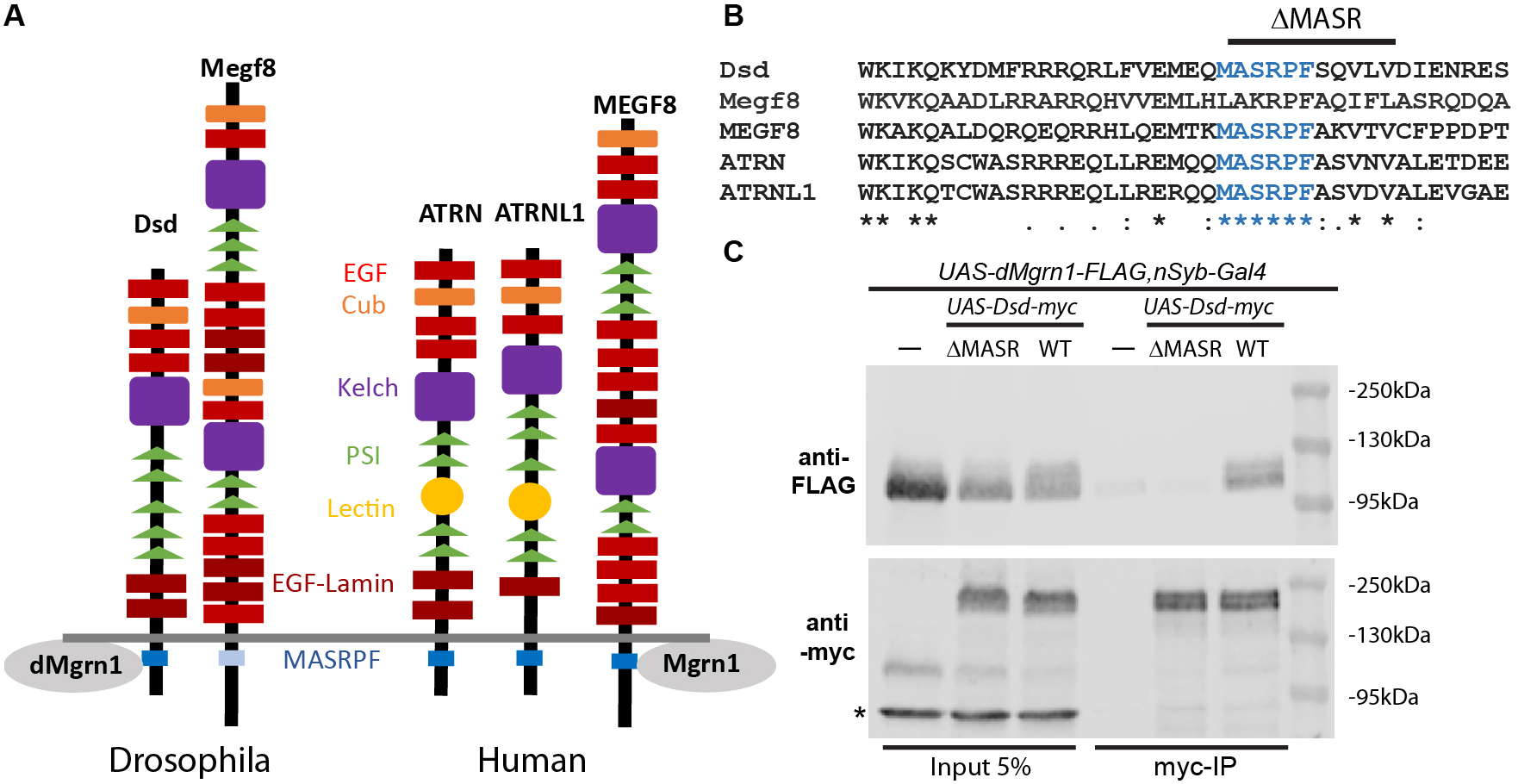
In vertebrates, the Attractin (Atrn) transmembrane protein family is comprised of ATRN, Attractin-like 1 (ATRNL1) and Multiple epidermal growth factor-like domain 8 (MEGF8) proteins, which contain an extensive extracellular domain with numerous protein interaction motifs and a highly conserved intracellular MASRPF motif (Fig. 1A and B) (Walker et al., 2007). Common to all three paralogs is that they have a strong phenotypic overlap with E3-ligase Mahogunin ring finger 1 (Mgrn1), due to regulation of the same G-protein coupled receptors (GPCRs) as Mgrn1. For example, rodent Atrn and Mgrn1 negatively regulate signaling melanocortin receptors MCR1 and MCR4 by promoting their endolysosomal trafficking. Thus, loss of function mutations in Atrn or Mgrn1 suppress the pigmentation and obesity phenotypes of MCR1/4 antagonist gain of function mutants (Barsh et al., 2002; He et al., 2003; Nagle et al., 1999). Loss of function of Megf8 and Mgrn1 mutants exhibit common developmental phenotypes (Aune et al., 2008; Cota et al., 2006; Zhang et al., 2009) and both proteins were identified as negative regulators of the GPCR Smoothened in mice (Pusapati et al., 2018). Most recently it was shown in vitro that Mgrn1 and Megf8 co-immunoprecipitate in a MASRPF motif-dependent manner, and this association is required for Mgrn1 to mediate membrane-tethered ubiquitination to modulate the signaling strength of Hedgehog morphogens in mice (Kong et al., 2020).
Distracted (dsd) is the sole Drosophila ortholog of human ATRN and ATRNL1 (Fig. 1A), while Drosophila Megf8, like rodent Megf8, is required for early development (Lloyd et al., 2018). In contrast to Drosophila Megf8, the MASRPF motif is fully conserved in Dsd (Fig. 1B). The DRSC Integrative Ortholog Prediction Tool uses 15 different algorithms to identify orthologous genes (https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl, Version 8.0, August 2019). For CG9941 (hereafter referred to as dMrgn1) more algorithms predicted that it is orthologous to mammal MGRN1 (human 11/15, rat 7/13, mouse 10/15) than for RNF157 (Ring Finger Protein 157; human 10/14, rat 4/15, mouse 10/15). While the zinc finger domain is highly conserved (83% amino acid identity) in dMgrn1 when compared to either mammal MGRN1 or RNF157 proteins, the entire protein is less well preserved (amino acid identity 35-37%/similarity 48-51%) in part due to the Drosophila protein being larger by approximately 210 amino acids when compared to mammalian Mgrn1.
Here, we generated myc-tagged Dsd containing (UAS-Dsd-myc) or lacking the MASRPF motif (UAS-DsdΔMASR-myc, Fig. 1B), and FLAG-tagged dMgrn1 (UAS-dMgrn1-FLAG) transgenic lines to determine whether Dsd and dMgrn1 are in a complex together in vivo, and if the interaction is dependent on the MASRPF motif. We panneuronally (nSyb-Gal4) expressed dMgrn1-FLAG alone or together with Dsd-myc or DsdΔMASR-myc, and used Drosophila heads to perform immunoprecipitation (IP) with myc-trap agarose beads. In western blots of fly head supernatant prior to IP, dMgrn1-FLAG is detected at 110 kDa and Dsd-myc at 160kDa (Fig. 1, lane 1-3) confirming protein expression. Multiple independent IP experiments were performed using two distinct transgenic lines for expression of Dsd-myc and DsdΔMASR-myc, while using the same transgenic line for expression of dMgrn1-FLAG. The results demonstrated that Dsd and dMgrn1 co-IP, suggesting they interact in vivo (n=4), and that this interaction was indeed dependent on the MASRPF motif (n=3, Fig. 1C). After co-IP, dMgrn1-FLAG protein was detected abundantly when it was co-expressed with Dsd-myc (Fig. 1C, lane 6) but absent or at similarly low levels as the negative controls (UAS-dMgrn1-FLAG,nSyb-Gal4, Fig. 1C, lane 4) when co-expressed with DsdΔMASR-myc (Fig. 1C, lane 5). These results imply that Drosophila is a valid model to study ATRN’s proposed role as a substrate adaptor for MGRN1 (Kong et al., 2020). Loss of function of Atrn and Mgrn1 in rodents is associated with mitochondrial dysfunction, elevated oxidative stress, and adult-onset spongiform neurodegeneration (Bronson et al., 2001; Cota et al., 2008; He et al., 2003; Kuramoto et al., 2001; Li et al., 2014; Muto and Sato, 2003; Paz et al., 2007). In humans, ATRN is a candidate gene for sporadic amyotrophic lateral sclerosis (Morahan et al., 2009), ATRN plasma levels are altered in early-onset Alzheimer’s disease (AD) patients prior to symptoms (Muenchhoff et al., 2016), and its gene expression is reduced in Parkinson disease (PD) patients (Glaab and Schneider, 2015). Since the ATRN/MGRN1 receptor substrates involved in nervous system survival have not been identified, future studies in Drosophila may provide insight into the relevant signaling pathways.
Methods
Request a detailed protocolCloning and mutagenesis of Dsd and dMgrn1 transgenes
All primers were generated by Integrated DNA Technologies (IDT, Iowa, USA). The Dsd open reading frame (ORF) without the stop codons was amplified with forward (CAC CAT GTC CCT GTT GCC ACC G) and reverse (TGT ACA ACT GTC TGG GTG CTG GCT CTG) primers from the LD14047 cDNA clone (Stock Number: 1301485, Drosophila Genomics Resource Center, Indiana, USA) and cloned into the pENTR/D-TOPO (Invitrogen, Massachusetts, USA). The dMgrn1 open reading frame without the stop codons was amplified with forward (ATG GGC AAC TTG TGA GCA G) and reverse (AAC ATT GAC GGC ATT CTG) primers from the LP12254 cDNA clone (Stock Number: 1344980, Drosophila Genomics Resource Center, Indiana, USA) and cloned into the pENTR/D-TOPO . In addition, the highly conserved motif MASRPFSQVLV (amino acids 1208-1218) was deleted in pENTR/D-TOPO-DSD entry clone with QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, California, USA) using forward (CGT CGA AAT GGA ACA GGA TAT CGA GAA CCG CG) and reverse (CGC GGT TCT CGA TAT CCT GTT CCA TTT CGA CG) primers. The vectors pENTR/D-TOPO containing Mgrn1, Dsd without or with the MASRPFSQVLV deletion (referred to as DsdΔMASR) were verified using restriction enzyme digests and Sanger sequencing (Genewiz, New Jersey, USA). Both Dsd entry vectors were recombined with the pTWM destination vector (Stock Number: 1107, Drosophila Genomics Resource Center, Indiana, USA) and the dMgrn1 entry vector with the pTWF destination vector (Stock Number: 1116, Drosophila Genomics Resource Center, Indiana, USA) using LR clonase. The recombination for the presence of six in-frame myc-codons for the Dsd constructs and the three in-frame FLAG codons for the Mgrn1 construct at the C-termini was verified using restriction enzyme digests and Sanger sequencing (Genewiz, New Jersey, USA). The multiple UAS-transgenic lines with P-element insertions at random sites on the second and third chromosome for all three constructs were established in the w1118 background by BestGene Inc (California, USA).
Fly strains and maintenance
The nSyb-Gal4 line (Bloomington Stock Center, Indiana, USA, RRID: BDSC_51635) was recombined with the UAS-dMgrn1-FLAG line to established a stable stock expressing dMgrn1-FLAG panneuronally. The UAS-dMgrn1-FLAG,nSyb-Gal4 stock was crossed to two distinct UAS-Dsd-myc or the UAS-DsdΔMASR-myc lines and the offspring were collected for the co-IP experiments. The UAS-dMgrn1-FLAG,nSyb-Gal4 stock was used as a negative control. All flies were reared on standard fly media at 25°C.
Co- immunoprecipitation
Collected adult flies (~3ml) were snap-frozen in liquid nitrogen in 15ml conical tubes, heads were severed by vigorous shaking, and collected by passing through a sieve. Samples were kept at 4oC for the rest of the experiment. 100mg of heads were homogenized in 450μl of lysis buffer (10 mM Tris base, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 10µM MG132, 1 mM PMSF, 20mM NEM, 1x Complete ULTRA EDTA-free Protease Inhibitor cocktail, 1xPhosStop) using a handheld homogenizer. Homogenates were centrifuged at 500xg for 1min to remove cuticle particulates, and the supernatant was collected and centrifuged again at 16000xg for 30min. 5% of the supernatant was saved as inputs while the rest of the supernatant was precleared with 60μl of Protein A agarose beads (Pierce™ Protein A Agarose, ThermoFisher Scientific, Massachusetts, USA) for 10min. Protein A agarose beads were removed by centrifugation at 2500xg for 2 min and supernatant immunoprecipitated with 25μl of Myc-Trap® (ChromoTek, Bavaria, Germany) agarose beads for 1hr. Beads were collected by centrifugation at 2500xg for 2 min, washed 3 times with 500μldilution buffer (10mM Tris base, 150mM NaCl), resuspended in 30μl 2x Laemmli buffer, boiled for 10 min at 95°C and pelleted at 2500xg for 2 min. Proteins in the supernatant were separated on an 8% SDS-PAGE gel. Western blots of the gels were first probed with mouse anti-FLAG® M2 (Sigma-Aldrich, Missouri, USA, RRID:AB_259529, dilution 1:1000) and with mouse anti-myc 9E11 antibody (BioLegend, California, USA, RRID: AB_2565039, dilution 1:1000) consecutively. Goat anti-mouse IRDye® 680RD (LI-COR Biosciences, Nebraska, USA, RRID:AB_10956588, dilution 1:15000) was used to detect the primary antibodies. Images were acquired and analyzed using Image Studio from LI-COR Biosciences.
Acknowledgments
Destination vectors and cDNA clone were obtained from Drosophila Genomics Resource Center (NIH Grant 2P40OD010949) and the nSyb-Gal4 line from the Bloomington Drosophila Stock Center (Indiana, United States, NIH P40OD018537).
References
Funding
The project was supported by the Jupiter Life Sciences Initiative and the Brain Institute pilot grant at Florida Atlantic University and the National Institute for Neurological Disease and Stroke (R15NS090043)
Reviewed By
AnonymousHistory
Received: March 27, 2021Revision received: June 14, 2021
Accepted: June 15, 2021
Published: July 2, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Nawaratne, V; Kudumala, S; Kakad, PP; Godenschwege, TA (2021). The conserved MASRPF motif in the Attractin homolog, Distracted, is required for association with Drosophila E3-ligase Mgrn1. microPublication Biology. 10.17912/micropub.biology.000416.Download: RIS BibTeX