Illinois State University, Normal, IL USA
Frostburg State University, Frostburg, MD USA
Nevada State College, Henderson, NV USA
University of Detroit Mercy, Detroit, MI USA
Abstract
Genetic screens are used to identify genes involved in specific biological processes. An EMS mutagenesis screen in Drosophila melanogaster identified growth control phenotypes in the developing eye. One mutant line from this screen, H.3.2, was phenotypically characterized using the FLP/FRT system and genetically mapped by complementation analysis and genomic sequencing by undergraduate students participating in the multi-institution Fly-CURE consortium. H.3.2 was found to have a nonsense mutation in short stop (shot), an ortholog of the mammalian spectraplakin dystonin (DST). shot and DST are involved in cytoskeletal organization and play roles during cell growth and proliferation.
Description
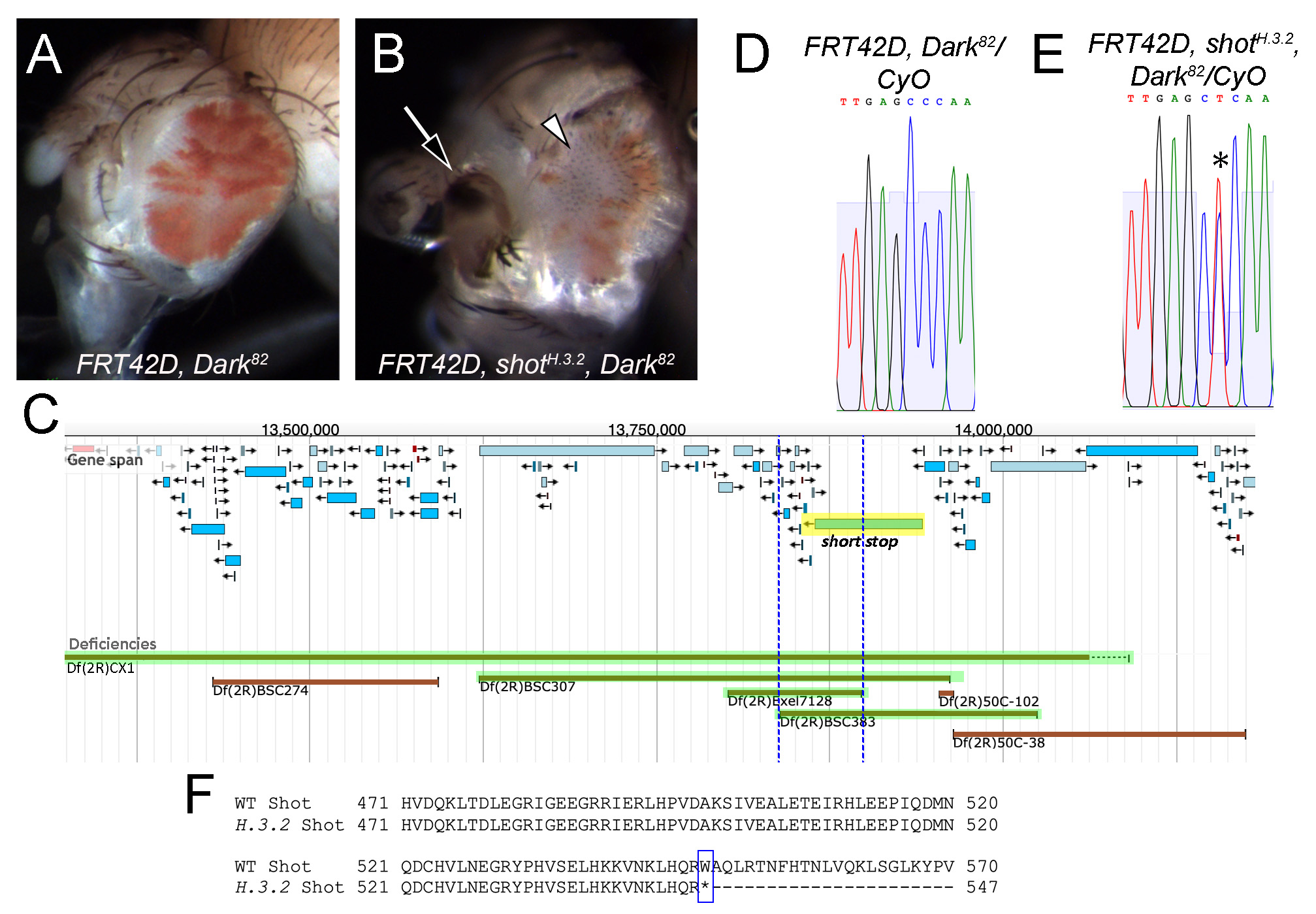
The homozygous lethal H.3.2 mutation was generated from an EMS mutagenesis screen utilizing the FLP/FRT mitotic recombination system on chromosome 2R to detect phenotypic abnormalities consistent with aberrant cell growth control in the developing eye (Kagey et al. 2012). Apoptosis was prevented using an allele of Death-associated Apaf-related killer (Dark; allele Dark82), which carries a mini-white transgene (w+mC) and allows for mutant cells to be identified by the presence of red pigmentation after mitotic recombination (Akdemir et al. 2006).
The phenotypic analysis and genetic mapping of mutant H.3.2 were carried out by independent groups of undergraduate researchers in genetics laboratory courses at three different institutions as part of the Fly-CURE consortium (Bieser et al. 2018). First, phenotypic analysis of H.3.2 was performed by crossing flies expressing the site-specific recombinase FLP under the control of a promoter for eyeless (ey) and FLP target sites on chromosome 2R (FRT42D) with flies carrying the H.3.2 mutation (genotype FRT42D, H.3.2, Dark82/CyO) or control flies (genotype FRT42D, Dark82/CyO). Mosaic eyes of the resulting progeny were quantified, showing that the H.3.2 mutation resulted in mosaic eyes with less mutant tissue (17% on average; Figure 1B) than control eyes (49% on average; Figure 1A). In addition, disorganized patterning of the ommatidia resulting in a rough eye phenotype, antennal defects, and other morphological defects in the tissue surrounding the eye were observed in H.3.2 mutant eyes (Figure 1B), but not in control eyes (Figure 1A). These data suggest that shot may be a conditional suppressor of proliferation non-cell autonomously and/or may promote proliferation cell autonomously.
Genetic complementation mapping of the H.3.2 mutation was performed by crossing FRT42D, H.3.2, Dark82/CyO females with males from each of the stocks in the Bloomington 2R Deficiency Kit (Cook et al. 2012). At least fifty progeny from each cross were evaluated for complementation based on lethality or survival of the homozygous lethal H.3.2 mutation in trans to each deficiency chromosome. Chromosomal breakpoints were obtained from the Bloomington Drosophila Stock Center for further analysis (Table 1). H.3.2 failed to complement deficiency lines Df(2R)CX1/SM1, Df(2R)BSC307/CyO, and Df(2R)BSC383/CyO (Table 1, Figure 1C). Additional crosses were performed with smaller deficiencies within the region (Table 1), allowing for a more refined region of overlap to 2R:13,839,479..13,897,827 (Figure 1C). Alleles of the gene short stop (shot), one of the 12 protein-coding genes in the region, failed to complement the H.3.2 mutation (Table 1).
Table 1. Complementation results with 2R deficiency lines and individual alleles of shot with FRT42D, H.3.2, Dark82. Deficiency mapping narrowed down the region of the H.3.2 mutation to 2R:13,839,479..13,897,827, which contains the gene short stop (shot).
| Bloomington 2R Deficiency Kit | |||
| Deficiency | BDSC Stock # | Region | Complementation Result |
| Df(2R)CX1/SM1 | 442 | 2R:12,700,421..14,091,140 | Fail to complement |
| Df(2R)BSC274/CyO | 23170 | 2R:13,430,464..13,593,272 | Complement |
| Df(2R)BSC307/CyO | 23690 | 2R:13,623,008..13,961,601 | Fail to complement |
| Df(2R)BSC383/CyO | 24407 | 2R:13,839,479..14,024,879 | Fail to complement |
| Df(2R)50C-38/CyO | 8114 | 2R:13,964,325..14,175,325 | Complement |
| Additional Deficiency Lines | |||
| Deficiency | BDSC Stock # | Region | Complementation Result |
| Df(2R)Exel7128/CyO | 7873 | 2R:13,801,956..13,897,827 | Fail to complement |
| Df(2R)50C-102/CyO | 8111 | 2R:13,954,125..13,964,325 | Complement |
| Individual Gene Alleles | |||
| Genotype | BDSC Stock # | Gene of interest | Complementation Result |
| b,1 wbSF20, Adhn4, shotSF20/CyO | 29033 | shot | Fail to complement |
| FRT42D, shotV104/SM5 | 8740 | shot | Fail to complement |
Because complementation mapping indicated candidate gene shot as a potential location for the H.3.2 mutation, genomic DNA was isolated from FRT42D, H.3.2, Dark82/CyO mutant and FRT42D, Dark82/CyO control flies for PCR and sequence analysis. Primers were designed to amplify exons of the shot gene and obtained from Integrated DNA Technologies. PCR products were purified and sequenced by Eurofins Genomics, Inc. to determine the molecular nature of the H.3.2 mutation.
Sequence analysis revealed a nucleotide change at 2R:13,904,428, corresponding to shot, in H.3.2 heterozygous flies. Where the control sequence showed a single C peak at this position (Figure 1D), the H.3.2 mutant sequence showed two peaks of T and C (Figure 1E). This change results in a nonsense mutation, wherein a tryptophan is mutated to a premature stop codon in all 21 of the annotated polypeptides encoded by the shot gene. This corresponds to a Trp548* change in a predicted spectrin-like repeat of isoform H, the longest annotated Shot polypeptide containing 8805 amino acids (Figure 1F). Therefore, this mutation results in Shot loss-of-function and the observed H.3.2 mutant phenotype. The increase of wildtype (white) cells in H.3.2 mutant mosaic tissue suggests a possible non-cell autonomous role of shot in eye development, wherein mutant cells signal to neighboring wildtype cells, promoting their proliferation. It is also possible that shot promotes cell proliferation autonomously (Su 2015). Additional experimentation is needed to better elucidate the molecular roles of shot during eye development.
Based on genetic mapping and sequence analysis in Drosophila melanogaster, we conclude that H.3.2 is a novel mutant allele of shot (shotH.3.2), resulting in a truncated protein. shot encodes a spectraplakin that interacts with microtubules, scaffold proteins, and certain signaling factors, and has been implicated in the development and maintenance of various tissues, including the nervous system, epidermis, and wing (Ricolo and Araujo 2020). shot was recently shown to have a role in cell growth via control of mitotic spindle assembly and chromosome dynamics (Dewey et al. 2020). RNAi knockdown of shot in Drosophila epithelia results in DNA double-stranded breaks (DSBs) due to errors in chromosome segregation during mitosis. shot has also been shown to play a role in regulating certain signaling molecules that direct cell spreading by controlling the actin cytoskeleton, thereby indicating a potential role for shot in suppressing tumor growth (Jain et al. 2019). Disruption of these or other mechanisms may contribute to the H.3.2 mutant phenotype observed in the eye.
The mammalian ortholog of shot, dystonin (DST), encodes an adhesion junction protein active in epithelial, neuronal, and muscular tissue, and is implicated in cell adhesion, cytoskeleton organization, and cell migration (Künzli et al. 2016). In addition, while shot was recently shown to suppress the transcriptional regulator YAP via mechanical cues, DST was similarly shown to be a novel tumor suppressor in the Hippo pathway (Jain et al. 2019). Therefore, future characterization of shotH.3.2 in Drosophila may contribute to evidence linking the cytoskeletal functions of these genes to growth control via cell autonomous and non-cell autonomous mechanisms.
Reagents
w–; FRT42D, Dark82/CyO (Akdemir et al. 2006)
w–; FRT42D, shotH.3.2, Dark82/CyO (this study)
w–, ey-FLP; FRT42D (BDSC 5616)
Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al. 2012)
Df(2R)Exel7128/CyO (BDSC 7873)
Df(2R)50C-102/CyO (BDSC 8111)
b1, wbSF20, Adhn4, shotSF20/CyO (BDSC 29033)
FRT42D, shotV104/SM5 (BDSC 8740)
shot forward primer: 5’ CCAACTTGTTTTGGCACCACTC 3’
shot reverse primer: 5’ CGAGGTTATCCTTCAGCAGG 3’
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
References
Funding
A. Vrailas-Mortimer is funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R15AR070505. Fly-CURE (K. Bieser, J. Kagey, J. Stamm, and A. Vrailas-Mortimer) is funded by a National Science Foundation IUSE Award (NSF 2021146).
Reviewed By
AnonymousHistory
Received: June 22, 2021Revision received: July 1, 2021
Accepted: July 2, 2021
Published: July 13, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Talley, EM; Watts, CT; Aboyer, S; Adamson, MG; Akoto, HA; Altemus, H; Avella, PJ; Bailey, R; Bell, ER; Bell, KL; Breneman, K; Burkhart, JS; Chanley, LJ; Cook, SS; DesLaurier, MT; Dorsey, TR; Doyle, CJ; Egloff, ME; Fasawe, AS; Garcia, KK; Graves, NP; Gray, TK; Gustafson, EM; Hall, MJ; Hayes, JD; Holic, LJ; Jarvis, BA; Klos, PS; Kritzmire, S; Kuzovko, L; Lainez, E; McCoy, S; Mierendorf, JC; Neri, NA; Neville, CR; Osborn, K; Parker, K; Parks, ME; Peck, K; Pitt, R; Platta, ME; Powell, B; Rodriguez, K; Ruiz, C; Schaefer, MN; Shields, AB; Smiley, JB; Stauffer, B; Straub, D; Sweeney, JL; Termine, KM; Thomas, B; Toth, SD; Veile, TR; Walker, KS; Webster, PN; Woodard, BJ; Yoder, QL; Young, MK; Zeedyk, ML; Ziegler, LN; Bieser, KL; Puthoff, DP; Stamm, J; Vrailas-Mortimer, AD; Kagey, JD; Merkle, JA (2021). Genetic mapping and phenotypic analysis of shotH.3.2 in Drosophila melanogaster. microPublication Biology. 10.17912/micropub.biology.000418.Download: RIS BibTeX