Department of Molecular, Cell, and Developmental Biology, University of California – Santa Cruz, Santa Cruz, CA, USA
Abstract
Until recently, the only verified component of Fibrous Bodies (FBs) within Caenorhabditis elegans spermatocytes was the Major Sperm Protein (MSP), a nematode-specific cytoskeletal element. Earlier studies in the pig parasite Ascaris suum had identified accessory proteins that facilitate MSP polymerization and depolymerization within the pseudopod of crawling spermatozoa. In this study, we show that C. elegans homologs of the two Ascaris accessory proteins MFP1 and MFP2 co-localize with MSP in both the pseudopods of C. elegans sperm and the FBs of C. elegans spermatocytes.
Description
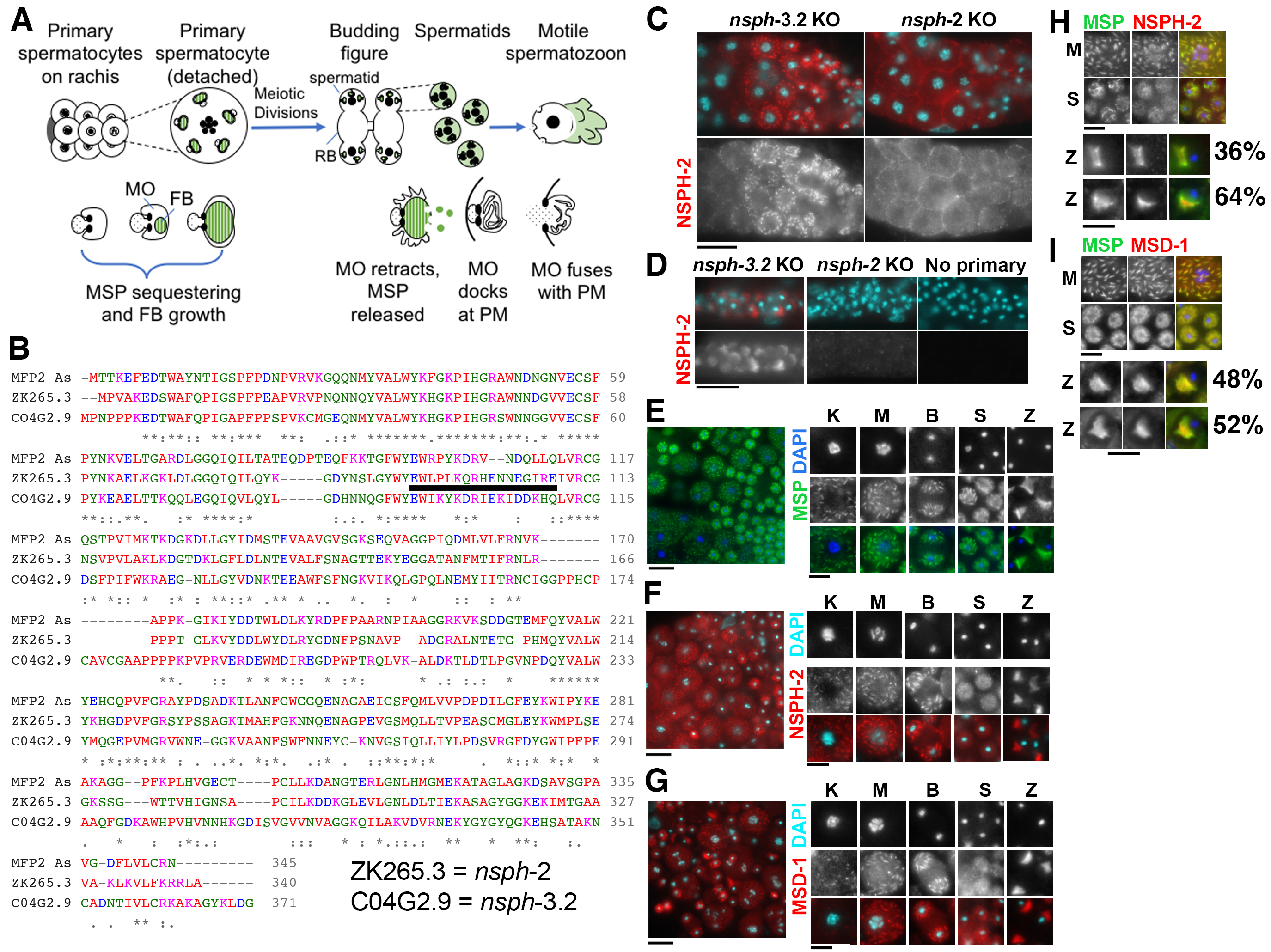
During the process of cell differentiation, specific cytoskeletal proteins can sequentially assemble into a wide variety of diverse molecular superstructures. Nematode spermatogenesis provides a powerful system for studying these transitions since sperm-specific transcription ceases prior to the meiotic divisions and translation ceases shortly thereafter (Chu and Shakes, 2013). Therefore, structural transitions that follow the meiotic divisions must be carried out by the remodeling of already synthesized proteins. The Major Sperm Protein (MSP) is a nematode-specific cytoskeletal element whose polymerization dynamics drive the pseudopod-based motility of the activated sperm (Roberts, 2005). In C. elegans, MSP additionally functions as the extracellular signaling molecule for triggering both ovulation and oocyte maturation (Miller et al., 2003). MSP is highly abundant in sperm, where it reaches 10-15% of total and 40% of soluble cellular protein (Roberts 2005). Within developing spermatocytes, MSP is packaged into fibrous body–membranous organelle (FB-MO) complexes (Fig. 1A, Roberts et al., 1986). By assembling into paracrystalline FBs, MSP is both sequestered away from the critical meiotic processes of chromosome segregation and cytokinesis while also being packaged for efficient segregation into spermatids during the post-meiotic partitioning process (Chu and Shakes 2013, Nishimura and L’Hernault, 2010, Price et al., 2021). Following the meiotic divisions and sperm individualization, FBs disassemble, and MSP disperses as dimers throughout the spermatid cytoplasm (Fig. 1A). When sperm activate to form motile spermatozoa, MSP polymerization within the pseudopod drives the motility of the crawling sperm (Chu and Shakes, 2013). Thus, MSP exists in at least three distinct molecular states: 1) in highly organized paracrystalline FBs within developing spermatocytes 2) as unpolymerized dimers within spermatids, and 3) in dynamically polymerizing filaments and fibers within crawling spermatozoa.
Because MSP neither binds nucleotides nor assembles into polar filaments, its assembly and disassembly dynamics require accessory proteins. Previous biochemical studies in the pig parasite Ascaris suum identified MSP fiber proteins 1 and 2 (MFP1/2) as key regulators of MSP polymerization (Sepsenwol et al., 1998, Buttery et al., 2003, Grant et al., 2005). MSP assembly dynamics within pseudopods requires the opposing forces of MFP1 and MFP2; MFP1 acts antagonistically to slow polymerization whereas phosphorylated MFP2 enhances MSP polymerization by serving as a nucleation site for polymer extension (Buttery et al., 2003, Grant et al., 2005). However, because these studies focused on MSP dynamics within crawling sperm and in vitro biochemical assays, the localization of these proteins within developing spermatocytes remained unknown. Would they be packaged along with MSP in FBs? Would the homologs of these proteins exhibit similar localization patterns during C. elegans spermatogenesis?
To confirm that the Ascaris proteins MFP2 and MFP1 have homologs in C. elegans, we compared protein sequences between the two species. We globally aligned Ascaris MFP2 with the predicted protein sequence of the two highest scoring C. elegans matches ZK265.3 and C04G2.9 (Fig. 1B). These are two spermatogenesis-expressed members of a larger MFP2 domain containing nematode specific peptide family, group H (Rödelsperger et al., 2021), and will be hereafter referred to by their C. elegans gene prefix nsph. Ascaris MFP2 exhibits 51.4% identity and 66.7% sequence similarity with ZK265.3 (nsph-2) and 38.0% identity and 52.3% similarity with C04G2.9 (nsph-3.2). ZK265.3 and C04G2.9 have 41.0% identity and 52.6% sequence similarity with each other. Ascaris MFP1 has two isoforms, MFP1-a and MFP1-b, which when aligned with C. elegans MSD-1 (Major Sperm protein Domain containing), show 52% identity and 70.8% sequence similarity, and 51.9% identity and 66.0% sequence similarity, respectively (Buttery et al., 2003). The sequence similarity between A. suum and C. elegans motility proteins suggests that they may have similar functions and binding properties.
To determine where NSPH-2 (MFP2) and MSD-1 (MFP1) localize within C. elegans spermatocytes and spermatozoa, we performed immunocytology experiments. For our analysis, we generated a rabbit polyclonal antibody against a unique NSPH-2 peptide (Fig. 1B) and obtained a polyclonal antibody against C. elegans MSD-1, from David Greenstein (Kosinski et al., 2005). To test the specificity of the new anti-NSPH-2 antibody, we used high-efficiency genome editing to generate CRISPR-Cas9 knockout (KO) lines through insertion of multiple premature termination codons in all three frames for both ZK265.3 (nsph-2) and CO4G2.9 (nsph-3.2) (Wang et al., 2018). Although neither knockout strain on its own exhibited a phenotype, they were useful for determining the specificity of our anti-NSPH-2 antibody. In control nsph-3.2 KO hermaphrodites, the anti-NSPH-2 antibody labelled distinct structures within late-stage spermatocytes (Fig. 1C) as well as the pseudopods of spermatozoa within the spermatheca (Fig. 1D). In nsph-2 KO hermaphrodites, only background labelling was observed. We then compared the localization of anti-NSPH-2 and anti-MSD-1 with the well characterized anti-MSP pattern in male gonads (Fig. 1E-G). In spermatozoa, both NSPH-2 and MSD-1 antibodies localize to the pseudopod, matching the known pattern in Ascaris spermatozoa. Within prophase and meiotically dividing spermatocytes, NSPH-2 and MSD-1 antibodies labelled discrete structures throughout the cytoplasm in a pattern strongly resembling FBs labelled by MSP antibodies (Fig. 1E-G). To verify that NSPH-2 and MSD-1 localized to FBs, we co-labelled male germlines with anti-MSP and either anti-MSD-1 or anti-NSPH-2 antibodies (Figure 1H-I). Within spermatocytes, the fully overlapping patterns revealed that NSPH-2 and MSD-1 are packaged along with MSP in the FBs. In spermatids, the MSP and MSD-1 were overlapping (Figure 1I). In contrast, except in immature spermatids that still retained their FBs, the NSPH-2 and MSP patterns only partially overlapped. In spermatids, the NSPH-2 labelling was notably less robust which may suggest that a change in protein conformation or binding partners is blocking the antigenic site. NSPH-2 does not appear to be degraded as it is present in the pseudopods of spermatozoa. All three antibodies labelled the pseudopods of spermatozoa, with the NSPH-2 and MSD-1 patterns either fully overlapping the MSP pattern (top set) or being restricted to a more central portion, adjacent to the cell body (bottom set).
Until recently, it was thought that Major Sperm Protein (MSP) was the only component of Fibrous Bodies (FBs) in C. elegans. Our discovery that MSD-1 and MFP2/NSPH-2 are packaged together with MSP in the FBs of developing spermatocytes supports a model in which the FBs function to gather, concentrate, and sequester proteins that will ultimately drive or regulate pseudopod motility. While early studies of MSP dynamics in Ascaris capitalized on its assets for biochemical approaches, parallel genetic approaches in C. elegans were stymied by the fact that many key factors, including MSP, MFP1 (MSD), and MFP2 (NSPH) are encoded by multigene families. Our own individual knockout strains of nsph-2 and nshp-3.2 exhibit no obvious phenotypes. This result does not mean that MFP2/NSPH is non-essential, but that exploring its function may require knocking out combinations of up to five of non-identical, sperm-related NSPH genes. Similarly, the four MSD genes encode identical proteins that may need to be deleted to generate a mutant phenotype. CRISPR technologies give us the ability to knockout multiple genes and explore MSP dynamics in new ways. Beyond MSD and NSPH, will other proteins first identified in Ascaris as regulators of MSP dynamics and sperm motility (Buttery et al., 2003) also localize to the FBs of developing spermatocytes? Alternatively, are there more proteins like the intrinsically disordered protein SPE-18 that localize to FBs and facilitate their growth and shaping but then are degraded shortly after the meiotic divisions (Price et al., 2021)? Further studies will reveal whether MSD-1 and NSPH-2 function together with SPE-18 to facilitate FB assembly, or if they are just conveniently packaged in FBs alongside MSP.
Methods
Request a detailed protocolWorm culture
Worms were cultured on MYOB plates (Church et al., 1995) and inoculated with the E. coli strain OP50, using methods similar to those described by Brenner (1974).
Creation of knock-out lines
CRISPR/Cas9 mutagenesis was performed as previously described (Paix et al., 2014; Dokshin et al., 2018; Wang et al., 2018). Briefly, C. elegans strain N2 was gene-edited by the insertion of a 43-base-pair sequence that includes multiple stop codons in all three reading frames to disrupt translation (Wang et al., 2018). A BamHI site was included 3’ to the stop codons to facilitate genotyping. Strains JDW307 nsph-3.2(wrd56[C04G2.9::exon 1 STOP]) and JDW308 nsph-2(wrd57[ZK265.3::exon 1 STOP]) were generated by CRISPR injection of an RNP complex [250 ng/µl of house-made Cas9 protein (Zuris et al., 2015) , 50 ng/µl of the relevant crRNA, and 100 ng/µl tracrRNA] that was first incubated at 37°C for 15 minutes, then mixed with the co-injection marker pRF4 (50 ng/µl) and the relevant repair oligos (110 ng/µl) before being injected into wild-type L4 + 1 day worms grown at 20°C. After 4 days, rolling worms were plated individually, allowed to lay eggs, and the parental animal was genotyped by PCR and BamHI digestion. For candidate knock-ins the extrachromosomal array was removed by selecting non-roller progeny. Genotyping primer sequences can be provided upon request. The tracrRNA and crRNAs were obtained from Integrated DNA Technologies (IDT).
C04G2.9 crRNA: 5’-TACCGGGCTCGGCGGGAAGG-3’
ZK265.3 crRNA: 5’-AAAGGAAAACTGGATTTGGG-3’
Repair oligos (BamHI sites in bold)
C04G2.9/NSPH-3.2 AAGATACTTGGGCATTCCAGCCAATCGGAGCCCCCTGGGAAGTTTGTCCAGAGCAGAGGTGACTAAGTGATAAGGATCCTCCCGCCGAGCCCGGTAAAATGTATGGGTGAGCA
ZK265.3/NSPH-2
TACAACAAAGCGGAGTTGAAAGGAAAACTGGATTTGGGAAGTTTGTCCAGAGCAGAGGTGACTAAGTGATAAGGATCCGGGTGGACAGATTCAAATTCTTCAGTACAAGGGA
Antibodies and immunocytochemistry
Sperm spreads were obtained by dissecting 8-15 male worms per slide in 7.5 microliters of egg buffer (Edgar, 1995) on ColorFrost Plus slides (Fisher Scientific, 12-550) coated with poly-L-lysine (Sigma Aldrich, P8290). Light pressure was applied to coverslips to flatten the samples. Samples were then freeze cracked in liquid nitrogen and fixed overnight in −20°C methanol. Specimen preparation and antibody labeling followed established protocols (Shakes et al., 2009). Primary antibodies included: 1:1500 4D5 mouse anti-MSP monoclonal (Kosinski et al., 2005) and 1:200 rabbit anti-MSD-1 rabbit polyclonal (Kosinski et al., 2005). Affinity purified rabbit antiserum against MFP2/NSPH-2 (ZK265.3) was generated by YenZym Antibodies using the peptide 92-108 EWLPLKQRHENNEGIRE. In experiments, the antibody was used at a 1:200 dilution. All samples were incubated with primary antibodies for 2 hours at room temperature. Affinity-purified secondary antibodies included 1:400 Alexa Fluor Plus 555 goat anti-rabbit IgG (Invitrogen, A32732) and 1:300 Alexa Fluor 488 goat anti-mouse IgG (H+L) (Jackson ImmunoResearch, 115-545-146). Final slides were mounted with Fluoro Gel with DABCO (Electron Microscopy Sciences #17985-02) containing DAPI.
Imaging and analysis
Images were acquired with epifluorescence using an Olympus BX60 microscope equipped with a QImaging EXi Aqua CCD camera. Photos were taken, merged, and exported for analysis using the program iVision. For control studies (Fig. 1C-D), image exposures were kept constant with no further image processing. For others, image exposures were optimized for individual gonads. The levels adjust function in Adobe Photoshop was used to spread the data containing regions of the image across the full range of tonalities.
Reagents
| Strain | Genotype | Available from |
| N2 | Wildtype C. elegans strain | CGC |
| CB4088 | him-5(e1490) V | CGC |
| JDW307 | nsph-3.2(wrd56[C0G2.9::exon 1 STOP]) | J. Ward lab |
| JDW308 | nsph-2(wrd57[ZK265.3::exon 1 STOP]) | J. Ward lab |
| Plasmid | Genotype | Description |
| pRF4 | co-injectable marker – dominant roller | 4 kb fragment of genomic DNA from rol-6(su1006) collagen gene in Bluescript vector (Mello et al., 1991). |
| Antibody | Animal and clonality | Description |
| anti-MSP | Mouse monoclonal, 4D5 | Kosinski et al., 2005 |
| anti-MSD-1 | Rabbit polyclonal, R194P | Kosinski et al., 2005 |
| anti-NSPH-2 | Rabbit polyclonal | Rabbit antiserum was generated by immunizing a rabbit against the aa 92-108 (EWLPLKQRHENNEGIRE) of C. elegans ZK265.3, YenZym |
| Alexa Fluor Plus 555, secondary | Goat anti-rabbit IgG | Invitrogen, A32732 |
| Alexa Fluor 488, secondary | Goat anti-mouse IgG | Jackson ImmunoResearch, 115-545-146 |
Acknowledgments
We would like David Greenstein for supplying antibodies against MSP and MSD-1.
References
Funding
The work was supported by a grant R15GM-096309 from the National Institutes of Health and the McLeod Tyler Professorship to D.C.S, a National Science Foundation (NSF) Division of Molecular and Cellular Biosciences CAREER award (1942922) to J.D.W., and an Honors Fellowship grant from the Charles Center at William & Mary to K.N.M.
Reviewed By
AnonymousHistory
Received: June 29, 2021Revision received: July 17, 2021
Accepted: July 19, 2021
Published: July 22, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Morrison, KN; Uyehara, CM; Ragle, JM; Ward, JD; Shakes, DC (2021). MFP1/MSD-1 and MFP2/NSPH-2 co-localize with MSP during C. elegans spermatogenesis. microPublication Biology. 10.17912/micropub.biology.000427.Download: RIS BibTeX