Department of Biology, Hastings College, Hastings, Nebraska 68901
Medical Mycology Research Center, Chiba University, Chiba 260-8673, Japan
Department of Materials Chemistry, National Institute of Technology (KOSEN), Asahikawa College, Asahikawa, Hokkaido 071-8142, Japan
Abstract
Autophagy is a conserved catabolic process by which eukaryotic cells respond to stress by targeting damaged or unneeded molecules or organelles for sequestration into specialized vesicles known as autophagosomes. Autophagosomes ultimately facilitate the digestion and recycling of their contents by fusing with the degradative organelle of the cell. Studies of the budding yeast Saccharomyces cerevisiae have revealed various types of stress that can regulate autophagy, including starvation and extreme temperatures. While autophagy has not yet been directly shown to confer the ability to survive extreme cold or freeze-thaw stress in yeast, upregulation of autophagy has been directly implicated in the ability of arctic insects to survive cold temperatures. We are interested in investigating the potential role of autophagy in polar habitat survival by cold-loving (psychrophilic) yeast like Mrakia blollopsis. To begin to examine the conservation of Atg machinery in polar-collected yeast, we focused on Atg8, a small, ubiquitin-like protein that plays an important role in autophagy. We report that Atg8 is conserved between S. cerevisiae and polar-collected yeast, using Atg8 from Mrakia blollopsis (strain TGK1-2) as an example. This study represents the first direct examination of autophagy machinery conservation across mesophilic and psychrophilic species of yeast.
Description
Autophagy is a stress response mechanism through which eukaryotic cells target and sequester unnecessary or damaged cellular components for degradation and recycling. The hallmark of the autophagy process is the formation of a large (600-900 nm in diameter), double-bilayered autophagosome, a temporary vesicle responsible for sequestering and transporting the cellular components to be recycled to the degradative organelle of the cell. The vacuole functions as the degradative organelle in yeast, where the molecular process of autophagy was first characterized. Autophagy is orchestrated by a group of proteins known as AuTophaGy-related or Atg proteins. Depending on the stress stimuli experienced, eukaryotic cells can deploy a number of different forms of autophagy. These include a general form known as macroautophagy that targets cellular components in bulk for degradation, as well as highly specific forms that target highly specific cellular components such as ribosomes (ribophagy), mitochondria (mitophagy), and peroxisomes (pexophagy). A specific set of core Atg protein machinery is shared among all of these forms of autophagy, and is essential for regulating and bringing about the formation of autophagosomes. Atg8p, coded by the ATG8 gene in Saccharomyces cerevisiae, is one of these core Atg proteins that is essential for autophagy in eukaryotes.
During autophagy, Atg8p undergoes an ubiquitin-like conjugation to the phospholipid phosphatidylethanolamine (PE) on the autophagosome membrane, where it helps recruit cargo and membrane material to facilitate its formation. Mature autophagosomes ultimately fuse with the vacuole, allowing for the recycling of their contents, including Atg8p itself, for degradation (Kirisako et al., 1999; Huang et al., 2000; Nair et al., 2012). In fact, Atg8 is considered as a standard autophagic marker that can be used to track progression of autophagy in cells at the microscopic and molecular levels (Klionsky et al., 2021).
Our understanding of autophagy as a cellular response has heavily relied on studies of the budding yeast Saccharomyces cerevisiae as a model system to reveal the molecular underpinnings of the process and the different types of stress that can regulate it, including starvation, exposure to reactive oxygen species, and extreme temperatures. Interestingly, autophagy has not yet been directly linked with the ability to survive extreme cold, despite the fact that freeze-thaw stress has been well characterized in laboratory strains of S. cerevisiae and other moderate temperature (mesophilic) yeast. On the other hand, upregulation of autophagy has been directly linked to the ability of an antarctic insect to survive cold temperatures (Teets et al., 2012; Teets and Denlinger, 2013). While autophagy has yet to be directly linked to polar habitat survival by cold-loving (psychrophilic) yeast like Mrakia blollopsis, a recent study identified the presence of stress-related genes in other species of psychrophilic yeast (Baeza et al., 2021).
To begin to examine the conservation of Atg machinery in polar-collected yeast, we set out to determine the degree to which the Atg8 protein is conserved between S. cerevisiae and species of polar-collected yeast such as Mrakia blollopsis (strain TGK1-2). This strain of Mrakia blollopsis was isolated from soil samples surrounding the Tokkuri Ike lake in the ice-free Skarvsnes area in East Antarctica (Tsuji et al., 2016; Tsuji et al., 2019). Atg8 was chosen in particular from a larger group of Atg proteins because of its central role in the autophagic cascade of molecular events. Like in S. cerevisiae, there appears to be only one ATG8 gene in M. blollopsis, strain TGK1-2.
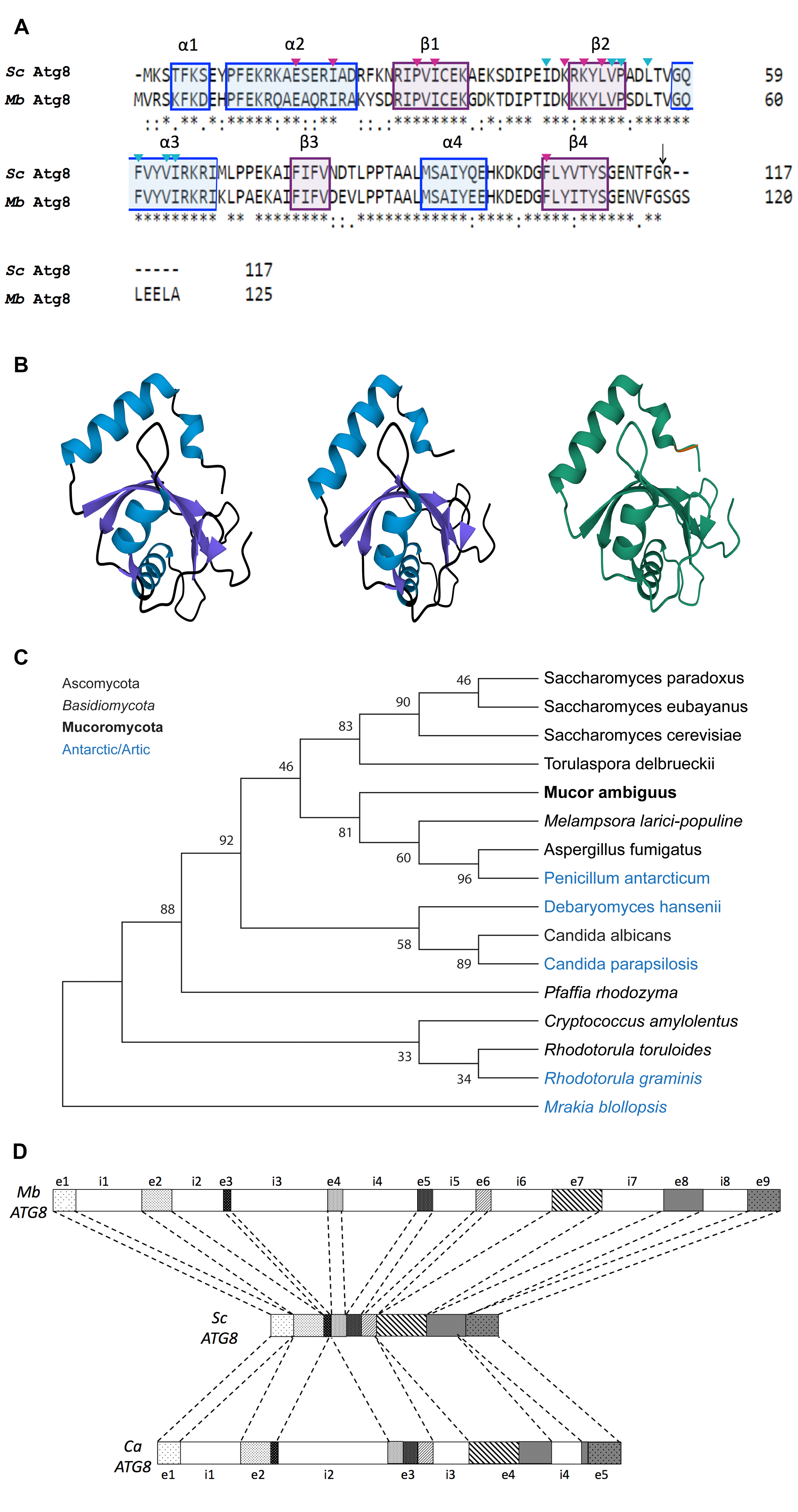
Protein sequence and structural alignment of Atg8p from Saccharomyces cerevisiae (Sc Atg8) and Atg8 from the psychrophilic yeast Mrakia blollopsis (strain TGK1-2; Mb Atg8) shows conservation of secondary structure domains and residues that have been identified as key for Atg8 function (Figure 1A and 1B). Conserved elements in Mb Atg8 that are characteristic of Atg8 proteins include the two amino-terminal α helices (α1 and α2) and a C-terminal core that structurally resembles ubiquitin (Noda et al., 2010; Shpilka et al., 2011; Wesch et al., 2020). As in Sc Atg8, the C-terminal tail of Mb Atg8 contains a conserved glycine residue, which is often referred to as a scissile or catalytic site (Figure 1A, indicated by an arrow). This glycine residue marks the cleavage site at which Atg8 can be conjugated to the membrane or lipidated (Noda et al., 2010; Shpilka et al., 2011; Klionsky and Schulman, 2014; Wesch et al., 2020). Similarly to that of Sc Atg8, the ubiquitin-like C-terminus of Mb Atg8 consists of a four-stranded β sheet and two α helices (α3 and α4) arranged in a β-grasp fold (Shpilka et al., 2011; Klionsky and Schulman, 2014; Wesch et al., 2020). It is likely that, as in Sc Atg8, Mb Atg8 serves as a scaffold for protein-protein interactions (Shpilka et al., 2011; Klionsky and Schulman, 2014; Wesch et al., 2020). For example, in characterized Atg8 proteins α1 and α2 align with the ubiquitin-like core, and α3 aligns with β2, to form two deep hydrophobic pockets (HP1 and HP2), respectively. HP1 and HP2 can in turn host protein-protein interactions to additional adaptor and scaffolding proteins (Wesch et al., 2020). Residues known to be important for HP1 and HP2 function in Sc Atg8, are also conserved in Mb Atg8 (Figure 1A). Given the conservation of key structural and functional elements, we hypothesize Mb Atg8 function is similar to that of Sc Atg8.
Construction of a maximum likelihood tree to highlight evolutionary relationships between Atg8 amino acid sequences from 16 fungal species (Figure 1C), including species of yeast known to inhabit antarctic and/or arctic environments, shows that the protein is well conserved (Figure 1A-B) despite the evolutionary distance between the Atg8 proteins from Mrakia blollopsis (strain TGK1-2) and other forms of cold-loving yeast (Figure 1C).
Furthermore, a search for conserved dinucleotides for splice site definition–GT at the 5’ splice site and AG at the 3’ splice site– revealed that the Mb (strain TGK1-2) ATG8 genomic sequence is likely a 9 exon/8 intron gene (Figure 1D). To assess differences in gene organization and Atg8 proteins across different phyla, Mb cold-adapted, mesophilic yeast (Sc), as well as fungi with higher intron densities than Sc (C. amylolentus), the ATG8 genomic, cDNA, and protein sequences of interest were compared. The M. blollopsis polypeptide (strain TGK1-2, 9 total exons) is 69% identical to S. cerevisiae and 77% identical to C. amylolentus. S. cerevisiae (strain ATCC 204508 / S288c, 0 exons) is 69% identical to M. blollopsis (strain TGK1-2) and 72% identical to C. amylolentus. C. amylolentus (CBS 6039, 5 total exons) is 72% identical to S. cerevisiae and 77% identical to M. blollopsis. The increased complexity in exon/intron gene organization from Sc (ascomycota-yeast) to Mb (basidiomycota-yeast) is synergistic with previous reports indicating that basidiomycetes such as Cryptococcus species display higher rates of alternative splicing (Grutzmann et al., 2014).
All in all, similarities between the Atg8 proteins from Saccharomyces cerevisiae and psychrophilic fungi indicate that it is conserved in these organisms. At the same time, some notable differences exist, such as the presence of splicing sites in the Mrakia blollopsis (strains TGK1-2) psychrophilic yeast gene. This study represents the first direct examination of autophagy machinery conservation across mesophilic and psychrophilic species of yeast. These findings will serve as a starting point for future investigations to ascertain the extent to which the function and regulation of these genes is conserved between mesophilic and psychrophilic yeast.
Methods
Request a detailed protocolSequencing: The genome of Mrakia blollopsis (strains TGK1-2) was sequenced using Pacific Biosciences (PacBio) Sequel. Gene prediction was carried out using the Funannotate pipeline.
Sequence and Structural Alignments: Clustal Omega (Sievers et al., 2011) was used to perform primary sequence alignments. For structural alignments, the pdb files of the Phyre2-predicted Sc Atg8p and Mb Atg8 protein structures (Kelley et al., 2015) were aligned using the topology-independent structure comparison algorithm CLICK (Nguyen et al., 2011). A hybrid pdb model was generated that highlights the consensus regions as well as the regions that are predicted to be different (Nguyen, et al., 2011). This hybrid pdb model was visualized using Mol* Viewer (Sehnal, et al., 2021). The CLICK algorithm was also used to obtain analysis parameters such as % structural conservation and root-mean-square deviation of atomic positions (RMSD). Splice sites were identified by aligning the genomic and cDNA sequences (Clustal Omega, EMBOSS Needle) with hand editing based on the presence of conserved splice site dinucleotide sequences (Sievers et al., 2011; Needleman and Wunsch, 1970).
Phylogenetic Analysis: The software MEGA was used to construct a maximum likelihood phylogeny with bootstrap support (500 iterations; Kumar et al., 2018). Individual species were chosen for this analysis to include Ascomycota, Basidiomycota and the basal Mucoromycota. Several polar-collected species were chosen to examine potential similarities between cold-adapted fungi from different divisions. Amino acid sequences of presumptive orthologues of the Atg8 protein from Mrakia blollopsis were obtained on Genbank (NCBI, 2016). The presumptive orthologue of Atg8 in Pfaffia rhodozyma was selected for phylogenetic comparison based on its closest match resulting from BLAST analysis (Altschul et al., 1990).
Acknowledgments
The authors would like to thank the Department of Biology and the Wanek School of Natural Sciences at High Point University (HPU) for resources and funding. Additional support was provided by HPU through Summer Undergraduate Research Program fellowships to BJI and HMS. HMS would like to thank the Natural Sciences Fellows Program at HPU for additional resources and mentorship.
References
Funding
This work was supported by JSPS KAKENHI Grant Number 16H06279 PAGS, JSPS Grant-in-Aid for Research Activity Start-up Grant (No. 15H06825) and Young Scientists (A) (No. 16H06211) to M. Tsuji. This study was also supported by National Institute of Polar Research (NIPR) through General Collaboration Project no. 2-29 to M. Tsuji. Additional support was provided by HPU through an HPU Research Advancement Grant to VAS (17-065) and Summer Undergraduate Research Program fellowships to BJI and HMS.
Reviewed By
AnonymousHistory
Received: July 26, 2021Revision received: August 9, 2021
Accepted: August 10, 2021
Published: August 16, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ivory, BJ; Smith, HM; Cabrera, E; Robinson, MR; Sparks, JT; Solem, A; Ishihara, Ji; Takahashi, H; Tsuji, M; Segarra, VA (2021). ATG8 is conserved between Saccharomyces cerevisiae and psychrophilic, polar-collected fungi. microPublication Biology. 10.17912/micropub.biology.000446.Download: RIS BibTeX