Department of Neuroscience, Cell Biology & Anatomy, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, Texas
Abstract
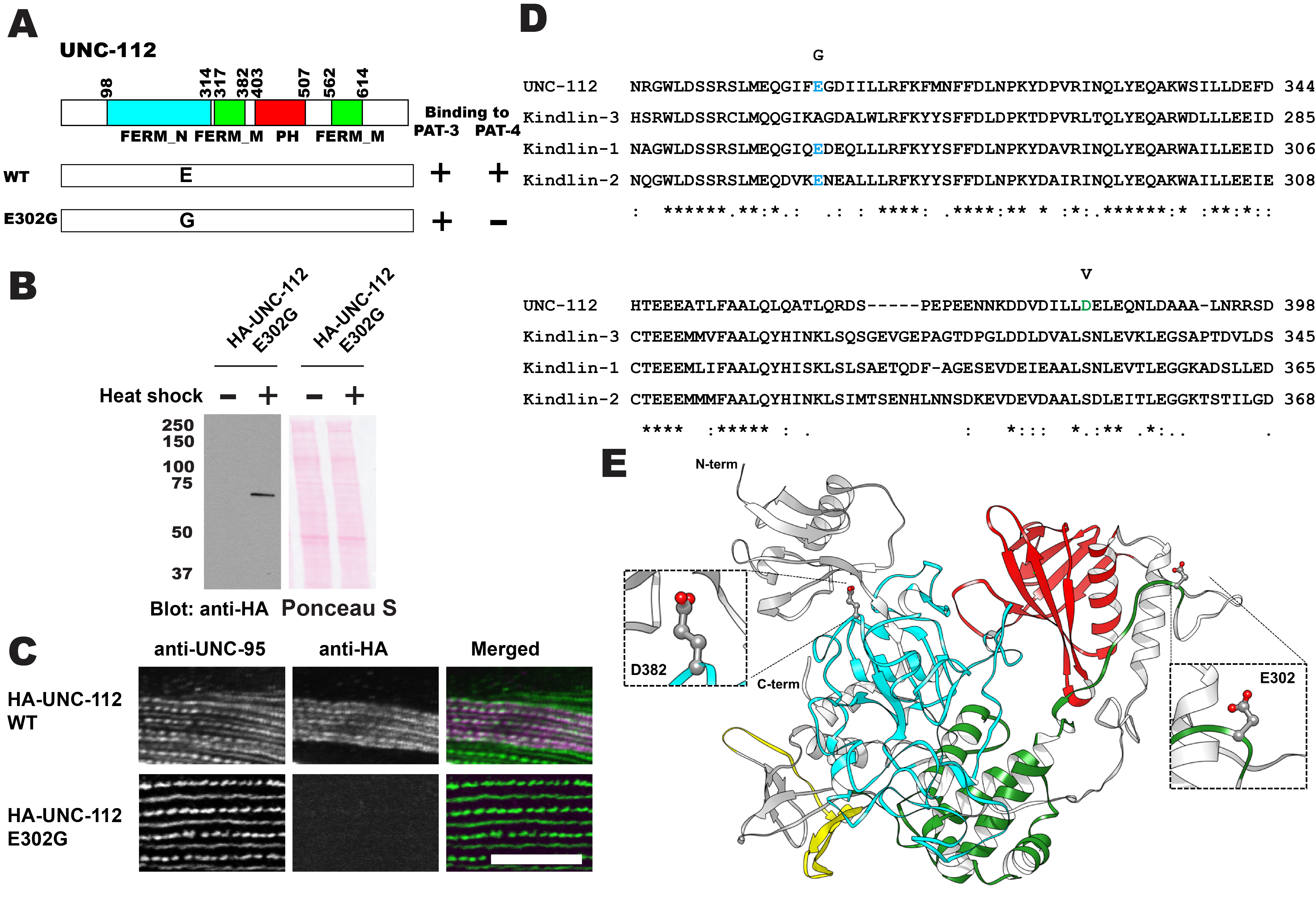
C. elegans UNC-112 (kindlin) is required for muscle sarcomere assembly, and is one component of a conserved four-protein complex that associates with the cytoplasmic tail of integrin at the base of integrin adhesion complexes in muscle. The four-protein complex consists of UNC-112 (kindlin), PAT-4 (integrin linked kinase; ILK), PAT-6 (alpha-parvin), and UNC-97 (PINCH). UNC-112 is comprised of 720 amino acid residues and contains FERM and PH domains. The N-terminal half of UNC-112 (1-396 aa) can bind to the C-terminal half of UNC-112 (397-720 aa), and this interaction is inhibited by the association of PAT-4 (ILK) to the N-terminal half of UNC-112. In support of this model, previously, we reported identification of a D382V mutation that results in lack of binding to PAT-4. However, this residue is not conserved in human Kindlins. Here, we report identification of a novel UNC-112 mutation of a conserved residue that cannot bind to PAT-4. UNC-112 E302G cannot bind to PAT-4 and does not localize to integrin adhesion complexes in muscle.
Description
C. elegans UNC-112 (kindlin) is required for muscle sarcomere assembly (Rogalski et al. 2000; Meves et al. 2009), and is one component of a conserved four-protein complex that associates with the cytoplasmic tail of integrin at the base of integrin adhesion complexes in muscle (Mackinnon et al. 2002; Lin et al. 2003; Norman et al. 2007; Qadota et al. 2014). UNC-112 binds directly with the cytoplasmic tail of PAT-3 (beta-integrin)(Qadota et al. 2012). UNC-112 binds to PAT-4 (ILK)(Mackinnon et al. 2002), and PAT-4 binds to both PAT-6 (alpha-parvin)(Lin et al. 2003), and UNC-97 (PINCH)(Mackinnon et al. 2002; Norman et al. 2007). A complex consisting of UNC-112, PAT-4, PAT-6 and UNC-97 has been demonstrated by co-immunoprecipitation (Qadota et al. 2014). UNC-112 is comprised of 720 amino acid residues and contains FERM and PH domains. The N-terminal half of UNC-112 (1-396 aa) can bind to the C-terminal half of UNC-112 (397-720 aa), and this interaction is inhibited by the association of PAT-4 (ILK) to the N-terminal half of UNC-112 (Qadota et al. 2012). In support of this model, we identified a D382V mutation that results in lack of binding to PAT-4 (Qadota et al. 2012). However, this residue is not conserved in human Kindlins (Figure 1D; D in UNC-112 but S in Kindlins), and mutation of this S to V in Kindlin-2 did not inhibit ILK binding (Huet-Calderwood et al. 2014). Here, we report identification of a novel UNC-112 mutation of a conserved residue that cannot bind to PAT-4. We found that E302G UNC-112 cannot bind to PAT-4, but still can bind to PAT-3 (beta-integrin) (Figure 1A). When we expressed HA tagged UNC-112 with E302G in C. elegans muscle (Figure 1B), HA tagged E302G UNC-112 cannot localize to the integrin adhesion complexes (dense bodies and M-lines) (Figure 1C). The E302 residue of UNC-112 is conserved in human Kindlin-1 and Kindlin-2, but not in Kindlin-3 (Figure 1D). Based on the only available crystal structure for a kindlin, that being for human kindlin-3 (Sun et al. 2020; Bu et al. 2020), we generated a homology model of UNC-112 and highlighted the locations of E302 and D382 (Figure 1E). Both E302 and D382 are located on surface loops of the structure. Mutating E302 to G, or mutating D382 to V resulted in no clashes with neighboring residues, based on the rotamer mutagenesis and energy minimization tools in Chimera (Pettersen et al., 2004). In fact, the lack of a sidechain of G, or the smaller sidechain of V, is likely to make these loops even more flexible. These mutations are predicted to not alter the overall structure of UNC-112 but could possibly affect the surface binding of UNC-112 to PAT-4. It should be noted, that in addition to our findings, a L located 6 residues C-terminal of D382, within the same FERM_M domain, and conserved in UNC-112 and all human kindlins (Figure 1D), when mutated to A, also greatly reduces binding to ILK (Huet-Calderwood et al. 2014). However, the results by Huet-Calderwood et al. were obtained by GST pulldown from lysates of tissue culture cells that overexpress both ILK and alpha-parvin. Our results, using the yeast two hybrid method and localization in worm muscle cells in which only UNC-112 was overexpressed, provide more evidence for an effect on direct binding between UNC-112 (kindlin) and PAT-4 (ILK). Our finding of a conserved residue in UNC-112 in the FERM_N domain that reduces binding to PAT-4, opens the door to testing the comparable residues in human Kindlin-1 and Kindlin-2 for their importance to binding to ILK. We do not know what the phenotype is of a nematode that is homozygous for the UNC-112 E302G mutation. Since UNC-112 function requires PAT-4 (Mackinnon et al. 2002), and all known mutations in pat-4 are Pat embryonic lethal, worms homozygous for UNC-112 E302G, might be Pat embryonic lethal. Several mutations in unc-112 are known to result in either the Pat embryonic phenotype or the adult viable Unc phenotype in which adults move slowly and have a disorganized myofilament lattice (Rogalski et al. 2000). The three known Pat alleles are unc-112(st562) and unc-112(st581) that are both nonsense mutations, and unc-112(gk1) that is a deletion. The Unc allele, unc-112(r367), is a missense mutation, T85I, and is also temperature sensitive. The molecular properties of UNC-112 T85I have not been explored, and thus, we do not know if the phenotype is due to reduced binding to PAT-4 or PAT-3, or whether it results in a unstable protein. Finally, there is one unusual allele created by CRISPR/Cas9, unc-112(kq715), L715E, which shows a defect in the migration of the distal tip cell (Park et al. 2020), but whether there was an effect on the myofilament lattice of body wall muscle, was not reported.
Methods
Request a detailed protocolYeast two-hybrid screening
Random mutagenesis using PCR and screening for interactions using the yeast two hybrid method was performed as previously described (Miller et al. 2006; Qadota et al. 2012). The UNC-112 N-terminal half was cloned into pGAD-C3 (prey plasmid), and the UNC-112 C-terminal half was cloned into pGBDU-C1 (bait plasmid). PAT-4 full length cDNA was cloned into pGBDU-C2 (bait plasmid). The N-terminal UNC-112 fragment was amplified by the error-prone method using the following primers: 5′ primer, AAA AAA GAG ATC GAA TTC CCC GGG GGA TCC; 3′ primer, GGT TTT TCA GTA TCT ACG ATT CAT AGA TCT. These primers were designed to amplify the insert, and each consists of 30 nucleotides of pGAD-C3. The cloning of error-prone PCR-amplified fragments into the acceptor plasmid was accomplished by exploiting yeast recombination in vivo. The mixture of the amplified PCR fragments (∼1 μg) and the acceptor plasmid (1 μg) digested with BamHI and BglII was transformed into PJ69-4A harboring pGBDU-UNC-112C. Transformed yeast cells were spread onto −Leu−Ura−His and 2 mM 3-amino-1,2,4-triazole to screen for His+ colonies. His+ selection ensured that the mutagenized UNC-112N could still interact with UNC-112C. This step was essential for eliminating clones with premature stop mutations or with many other mutations. His+ colonies were streaked onto an −Ade plate and screened for His+Ade+ colonies. After streaking on a 5-fluoroorotic acid plate to eliminate the URA3 marker bait plasmid (pGBDU-UNC-112C), prey clones were isolated from yeast and amplified in E. coli. From His+Ade+ yeast colonies, 64 mutagenized clones were isolated. These prey clones were transformed separately into PJ69-4A carrying either pGBDU-UNC-112C or pGBDU-PAT-4 (full-length) to check for interaction with the C-terminal half of UNC-112 and interaction with full-length PAT-4. Among 64 mutagenized clones of UNC-112N, 8 of these clones could not bind to PAT-4. From DNA sequencing of these 8 clones, we identified one clone with a single amino acid change, E302G. The UNC-112N with E302G was cloned into pACT-Q-UNC-112C (Qadota et al. 2012), resulting in pACT-Q-UNC-112 E302G (full length), and then used to test for binding to PAT-3.
Expression in C. elegans
To express HA-UNC-112 E302G in C. elegans using a heat shock promoter, the SmaI-EcoRV fragment of pACT-Q-UNC-112 E302G was inserted into EcoRV digested pKS-HA-UNC-112-Acp (Qadota et al. 2012), resulting in pKS-HA-UNC-112 E302G. The NheI fragment of pKS-HA-UNC-112 E302G was then cloned into NheI-digested pPD49.78 and pPD49.83, resulting in pPD49.78-HA-UNC-112 E302G and pPD49.83-HA-UNC-112 E302G. pPD49.78/83-HA-UNC-112 E302G were mixed with pTG96 (SUR-5::NLS::GFP) (Yochem et al., 1998) as a transformation marker and injected into wild type N2 worms. Transgenic lines with extrachromosomal arrays containing pPD49.78/83-HA-UNC-112 E302G and pTG96 (GB339; sfEx74 [sur-5::GFP; hspp::HA::unc-112 E302G]) were established by picking GFP-positive worms using a GFP dissection microscope. Generation and characterization of transgenic animals expressing by heat shock the comparable HA-tagged wild type UNC-112 was described previously (Qadota et al. 2012). Expression of the HA-tagged UNC-112 proteins (E302G and wild type) was induced by incubation of the transgenic worms at 30 °C for 2 h (heat shock).
Fluorescence imaging
Heat-shocked transgenic worms were fixed (Nonet et al. 1993) and stained by anti-GFP (to verify the existence of extrachromosomal array; rabbit polyclonal from Thermo Fisher A11122), anti-UNC-95 (to identify dense bodies and M-lines in muscle cells; rabbit polyclonal Benian-13, Qadota et al. 2007), and anti-HA (to determine the localization of HA-tagged UNC-112 proteins; Sigma-Aldrich H3663; 1:200 dilution). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat ×63/1.4 numerical aperture oil objective, in ×2.5 zoom mode. The color balances of the images were adjusted by using Adobe Photoshop version 22.4.3.
Western blotting
We prepared worm lysates (Hannak et al. 2002) from transgenic worms with or without heat shock and examined the expression of HA-tagged UNC-112 proteins by Western blot, reacting with anti-HA (Sigma-Aldrich H3663; 1:200 dilution).
Protein structure modeling
For UNC-112 protein structure modeling, CLUSTALW version 1.2.2 (https://www.ebi.ac.uk/Tools/msa/clustalw2/), SWISS-MODEL version July 2021 (https://swissmodel.expasy.org/; Waterhouse et al., 2018) and Phyre2 version 2.0 (http://www.sbg.bio.ic.ac.uk.phyre2/html/page.cgi?id=index; Kelley LA et al., 2015) online tools were used. Human kindlin-3 (7C3M.pdb; Bu et al. 2020) was used as reference crystal structure. Molecular graphics were generated by using Chimera version 1.15 (https://www.cgl.ucsf.edu/chimera/; Pettersen et al., 2004).
Acknowledgments
We thank Dr. Andrew Fire (Stanford University) for C. elegans expression vectors (pPD49.78 and pPD49.83).
References
Funding
This study was supported by a previous grant from the American Heart Association (11GRNT7820000).
Reviewed By
AnonymousHistory
Received: August 10, 2021Revision received: September 2, 2021
Accepted: September 7, 2021
Published: September 14, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Qadota, H; Oberhauser, AF; Benian, GM (2021). Missense mutation of a conserved residue in UNC-112 (kindlin) eliminates binding to PAT-4 (ILK). microPublication Biology. 10.17912/micropub.biology.000454.Download: RIS BibTeX