University of Michigan
Abstract
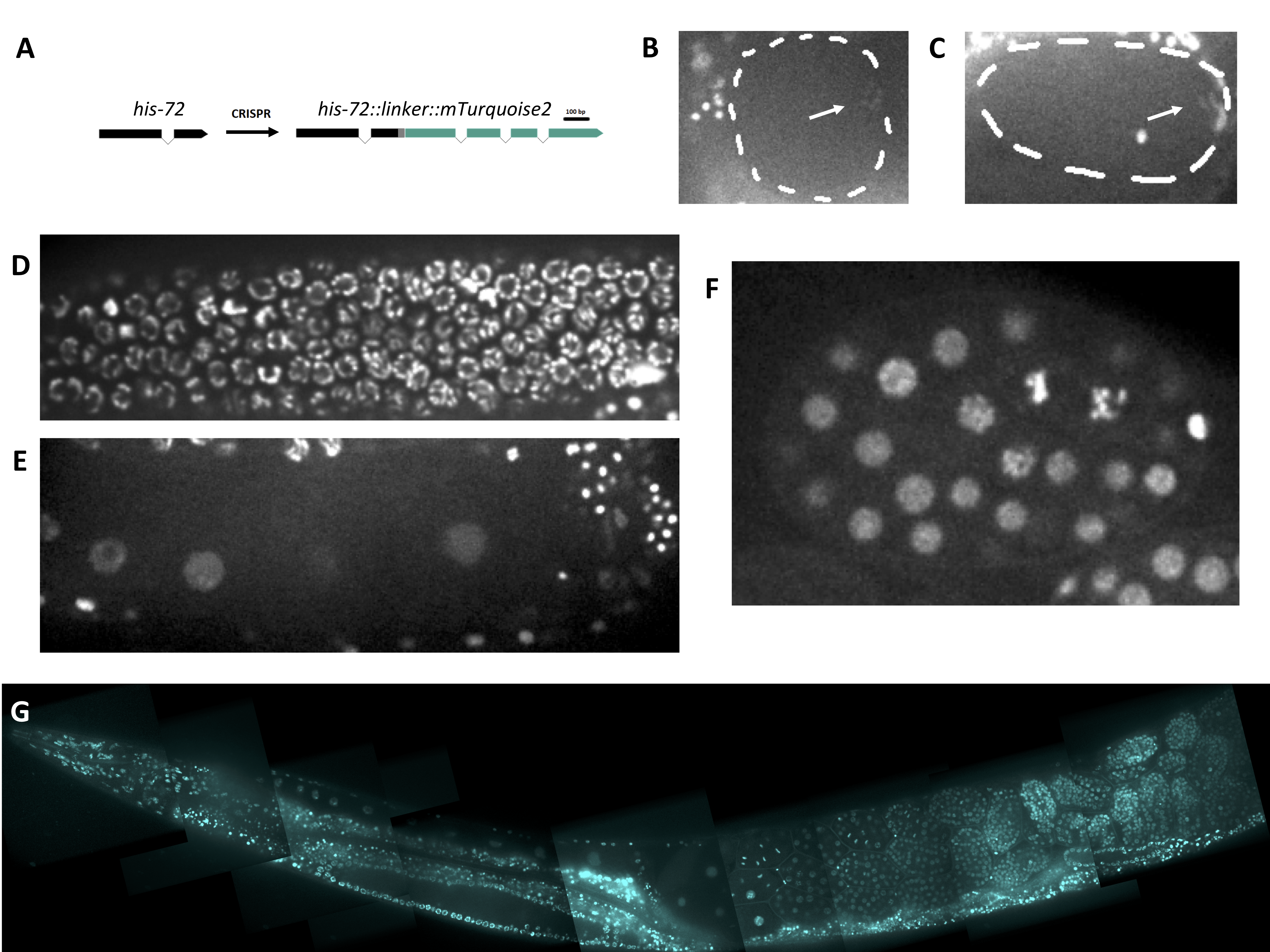
To generate a non-red/green fluorescent fusion histone protein in C. elegans, we have generated a C-terminal mTurquoise2-tagged HIS-72 at the endogenous locus using CRISPR. We found that HIS-72::mTurquoise2 localizes in a similar pattern to the previously published HIS-72::GFP strain.
Description
In fixed samples, chromatin is most often visualized by using a fluorescent DNA-binding dye like DAPI, which emits blue light (Kapuscinski 1995). In live fluorescence microscopy, however, a fluorescently tagged histone is often used instead. Histones form nucleosomes to package chromatin, and are involved in the organization of the genome (Chen et al. 2021). Most frequently, histones are fused to red or green fluorescent proteins, though the use of a blue or cyan fluorescent protein instead would allow for three (or more) channel imaging (Kanda et al. 1998, Das et al. 2003, Ooi et al. 2006).
We sought to generate an endogenously tagged histone line with a blue or cyan fluorescent protein. mTurquoise2 is a relatively recently engineered cyan fluorescent protein with a high quantum yield (Goedhart et al. 2012). Thus, we constructed a HIS-72::mTurquoise2 fluorescent fusion protein using a design similar to that of another group who also tagged HIS-72 (Figure 1A, Dickinson et al. 2013; see Design section). We report similar localization of HIS-72::mTurquoise2 to previously generated HIS-72 fluorescent fusion strains in most nuclei of the animal (Figure 1G). HIS-72::mTurquoise2 shows chromatin labeling in the germline and all stages of the cell cycle in the early embryo (Figure 1B-F). We observed that HIS-72::mTurquoise2 signal is significantly reduced on chromatin during the meiosis I and II divisions as compared with other stages (Figure 1B,C), which is not observed in other exogenously expressed fluorescently tagged histone lines (Bai and Bembenek 2017, Bembenek et al. 2007). This may reflect the role of different histone variants in meiotic chromosome function.
In sum, we have constructed a HIS-72::mTurquoise2 fluorescent fusion strain for use to the C. elegans community.
Methods
Request a detailed protocolCRISPR/Cas9 Gene editing
We followed the CRISPR/Cas9 protocol generated by the Seydoux lab for C-terminal mTurquoise2 tagging of the C. elegans his-72 gene (Paix et al. 2015). The repair template was amplified from the pDD377 plasmid, a gift from Bob Goldstein (Addgene plasmid #91823). Tagging of his-72 was done in the wild type N2 background. The transgenic strain was crossed to the N2 strain for 5 generations to remove potential off target mutations. All guide RNAs and oligos were obtained commercially.
The primer sequences for amplifying the repair template are listed below:
his-72::mTurquoise2_Forward (DS_030):
TGCAACTCGCCAGACGCATCAGAGGAGAACGTGCTGGAGCATCGGGAGCCTCAGGAGCATCGATGGTAAGTAAGGGCG
his-72::mTurquoise2_Reverse (DS_031):
ATTAAAAGTGCTTCGAGAATTGGTGATGGAGCTTACTTGTAGAGCTCGTCCATTCC
The flexible linker sequence: GGAGCATCGGGAGCCTCAGGAGCATCG (AA seq: GASGASGAS)
The target sequence (minus PAM) used in the crRNA (Termed DS_g004; see Dickinson et al., 2013): GAGCTTAAGCACGTTCTCCG
NOTE: This design only tags HIS-72 isoform a (Y49E10.6a, see WormBase).
Microscopy
Gravid adult worms were mounted on agar pads according to standard methods. Imaging was performed on a spinning disk confocal system consisting of a Nikon Eclipse inverted microscope with a 60×1.40 NA objective, a CSU-22 spinning disk system, and a Photometrics EM-CCD camera from Visitech International. Images were acquired by Metamorph (Molecular Devices) and analyzed by ImageJ/Fiji Bio-Formats plugins (National Institutes of Health) (Schindelin et al. 2012). The cyan/blue channel included a 450/50 emission filter with maximum blocking at 405 nm, and maximum intensity from 425-475 nm.
Reagents
JAB191: his-72(erb77[his-72::linker::mTurquoise2])
This strain will be made available to the CGC.
Acknowledgments
We thank members of the Bembenek lab, especially Chris Turpin, for useful feedback.
References
Funding
National Institutes of Health (R01 GM114471)
Reviewed By
AnonymousHistory
Received: August 24, 2021Revision received: September 11, 2021
Accepted: September 13, 2021
Published: September 29, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Sloan, DE; Bembenek, JN (2021). Endogenous expression and localization of HIS-72::mTurquoise2 in C. elegans. microPublication Biology. 10.17912/micropub.biology.000471.Download: RIS BibTeX