Abstract
LIN-41 (TRIM71), an ancient protein best known for its role in timing mitotic stem cell lineages, has been recently shown to be involved in postmitotic neurons to time their differentiation and post-differentiation. Here, we report the identification of 276 LIN-41 protein-expressing neurons in the C. elegans nervous system by NeuroPAL and a CRISPR-engineered mNG::LIN-41 reporter, which represents 91% of all hermaphrodite neurons and includes 87 neurons that were not previously reported by CeNGEN using single-cell RNA-seq. Broad lin-41 protein expression in C. elegans neurons suggests a widespread role of LIN-41 (TRIM71) in timing neuronal assembly, plasticity, and maintenance.
Description
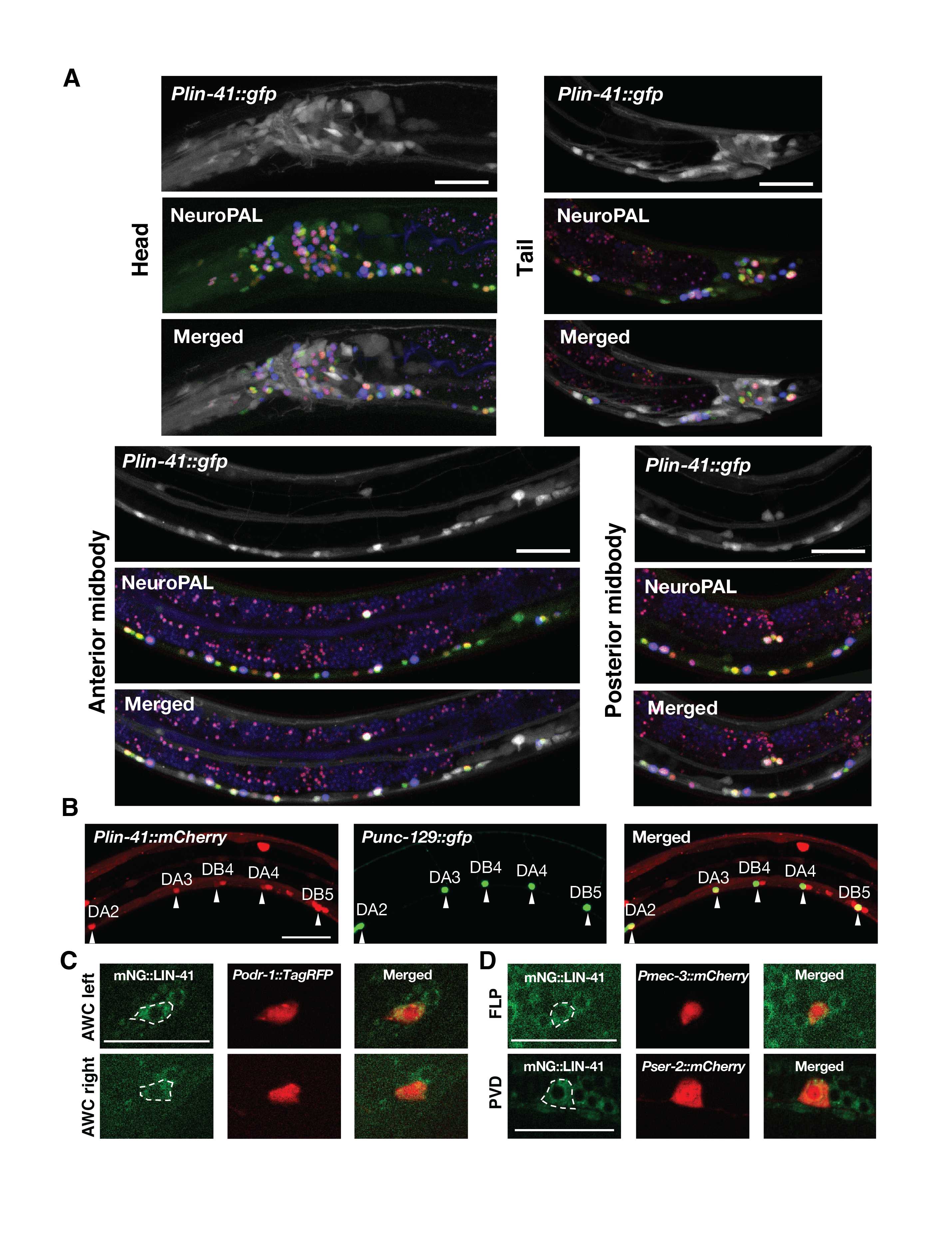
LIN-41 (TRIM71) proteins, which are evolutionarily conserved and best known for their roles in the timing of events in mitotic stem cell lineages, have been recently shown to be reutilized in postmitotic neurons to time differentiation and post-differentiation events. The importance of LIN-41 in the nervous system of C. elegans has begun to emerge, which includes temporal regulation of developmental decline in neuronal regeneration, sexually dimorphic nervous system differentiation, and sexual maturation of the male nervous system (Zou et al., 2013; Pereira et al., 2019; Lawson et al., 2019). To get a glimpse of how broadly lin-41 may be involved in the wiring and rewiring of the nervous system, it is important to first understand what neuron types are normally expressing LIN-41 proteins at the second and third larval stages during which LIN-41 protein expression peaks in the nervous system. In this study, we use the recently developed NeuroPAL technology for nervous-system-wide neuronal identification through whole-brain imaging (Yemini et al., 2021). Worms expressing NeuroPAL display a stereotypical multicolor fluorescence map for the entire hermaphrodite nervous system with unique color barcode created in each neuron, enabling identification of all neurons that also exhibit fluorescence of a reporter gene in the green emission channel. Here, we determine the brain-wide expression patterns of LIN-41 proteins by engineering a reporter strain with the mNeonGreen (mNG) Cassette tagged in the endogenous lin-41 gene using the CRISPR/Cas9 technology and co-labeling it with the NeuroPAL multicolor barcode. Although expression patterns of lin-41 and other genes in the nervous system were recently reported by the C. elegans Neuronal Gene Expression Map & Network (CeNGEN) project (Hammarlund et al., 2018; Taylor et al., 2021), our study provides complementary and further insights into lin-41-expressing neurons due to two important considerations. First, CeNGEN employs bulk RNA-sequencing of individual neuron classes from L4-stage larval animals to survey molecular mapping when neuronal development and connectivity are largely complete. In contrast, our study focuses on analyzing at L2- and L3-larval stages during which lin-41 expression peaks in the nervous system to maximize our ability to identify the lin-41-expressing neurons. Second, CeNGEN largely relies on cell sorting followed by RNA expression profiling, which might mask the protein expression of some genes in certain neurons where they undergo post-transcriptional gene regulation. For example, translation and stability of lin-41 mRNAs are known to be regulated by the let-7 microRNA. In contrast, our study reveals endogenous LIN-41 protein expression levels and their localization patterns in neurons brain-wide.
In summary, we have identified 276 LIN-41 protein-expressing neurons (Extended data, Table 1). Most of these LIN-41 protein-expressing neurons were also confirmed by a lin-41 promoter driving GFP reporter, which is under control by a constitutive unc-54 3’UTR (Figure 1). This consortium of neurons represents 91% of all hermaphrodite neurons and includes 87 neurons that were not previously reported by CeNGEN using single-cell RNA-seq (Extended data, Table 1; Taylor et al., 2021). For those 87 neurons that were identified as LIN-41 protein-expressing but not lin-41 mRNA-expressing (Extended data, Table 1), a possible explanation could be that a low-level lin-41 mRNA expression combined with a low-level let-7 microRNA-mediated translational repression could result in a detectable level of LIN-41 protein expression. In addition, the top 10 neurons identified by CeNGEN based on the level of lin-41 mRNA expression are not on the top 30 neuron list identified by NeuroPAL based on the frequency by which the mNG::LIN-41 fluorescence signal can be detected. The intensity of the mNG::LIN-41 fluorescence signal among different neurons is rather similar (Figure 1, C and D). Our results show that LIN-41 (TRIM71) proteins are broadly expressed in neurons, not just in the peripheral but also in the central nervous system (Figure 1; Extended data, Table 1), suggesting a widespread role of LIN-41 (TRIM71) in timing neuronal assembly, plasticity, and maintenance. (Zou et al., 2013; Chiu and Chang, 2013; Ivakhnitskaia et al., 2016; Ivakhnitskaia et al., 2017).
Methods
Request a detailed protocolStrains
C. elegans strains were cultured using standard methods (Brenner, 1974). All strains were grown at 20°C. Standard protocol was used for the strain constructions. Strains used in this study are listed below.
| XN2742 | lin-41(xr76)[mNG::LIN-41]I ; otIs670[NeuroPAL markers]V |
| XN2797 | otIs669[NeuroPAL markers]V; xrEx1151[Plin-41::gfp (50ng/ul)] |
| XN2803 | lin-41(xr76)I; vyIs56[Podr-1::TagRFP]III |
| XN2557 | lin-41(xr76)I; xrEx1010[Pmec-3::mCherry (5ng/ul)] |
| XN2540 | lin-41(xr76)I; xrEx997[Pser-2::mCherry (5ng/ul)] |
| XN2806 | evIs82b[Punc-129::gfp]IV; xrEx518[Plin-41::mCherry (50ng/ul)] |
Microscopy and NeuroPAL
Animals were mounted on 2% agarose pads and anesthetized with 7.5 mM Tetramisole. NeuroPAL images were taken in live animals using a 40x, 1.3 NA objective on a Zeiss LSM 880 confocal microscope, equipped with 7 laser lines: 405, 458, 488, 514, 561, 594, and 633 nm. Neuron types in the head and tail regions were annotated using NeuroPAL ID software. Midbody region neurons were manually annotated. The mNG::LIN-41 fluorescence intensity was analyzed by the NeuroPAL ID software. The linear change point was used as the threshold to determine LIN-41 protein-expressing neurons (Yemini et al., 2021). All other images were acquired using 40x, 1.4 NA oil objective on a Zeiss Axio M2 imager equipped with Apotome. All images were acquired at L2-L3 stages.
Generation of the mNG::LIN-41 knock-in using CRISPR-Cas9-triggered homologous recombination
The N-terminal mNG tagged lin-41(xr76)[mNG::LIN-41] allele was generated by CRISPR-Cas9 mediated genome editing using the self-excising cassette strategy (Dickinson et al., 2013; Dickinson et al., 2015). The lin-41 repair template homology arms and sgRNA were designed as previously described (Spike et al., 2014). The following mix was injected into N2 animals: the repair template: mNG^SEC^3xflag^lin-41 N term (50 ng/μl), the sgRNA and Cas9-expressing construct: Peft-3::Cas9::U6p::lin-41sgRNA-N-term (50 ng/μl), co-injection markers: Prab-3::mCherry (10 ng/μl), Pmyo-2::mCherry (2.5 ng/μl), and Pmyo-3::mCherry (5 ng/μl). The correct knock-in of the mNG marker was validated by PCR and sequencing. No noticeable phenotype, judged by normal morphology, fertility, behaviors, and growth rate, was observed in the lin-41(xr76)[mNG::LIN-41] animals.
Acknowledgments
This work was funded by grants from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM111320). We thank Dan Shaye and Jan K. Kitajewski for their generosity in sharing Zeiss LSM 880 confocal microscope system, Eunseo Kim for compiling the list of LIN-41 protein-expressing neurons, the reviewer for helpful comments, the Caenorhabditis Genetics Center for strains, and WormBase for readily accessible information.
Extended Data
Mushaine Shih, & Chieh Chang. (2021). List of LIN-41 protein-expressing neurons (Version 1.0) [Data set]. CaltechDATA. 10.22002/D1.2125
References
Funding
National Institute of General Medical Sciences of the National Institutes of Health (R01GM111320)
Reviewed By
Douglas PortmanHistory
Received: August 27, 2021Revision received: September 15, 2021
Accepted: September 21, 2021
Published: September 23, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Shih, M; Chang, C (2021). Brain-wide identification of LIN-41 (TRIM71) protein-expressing neurons by NeuroPAL. microPublication Biology. 10.17912/micropub.biology.000472.Download: RIS BibTeX