California State University Stanislaus, Turlock, CA USA
University of Evansville, Evansville, IN USA
Abstract
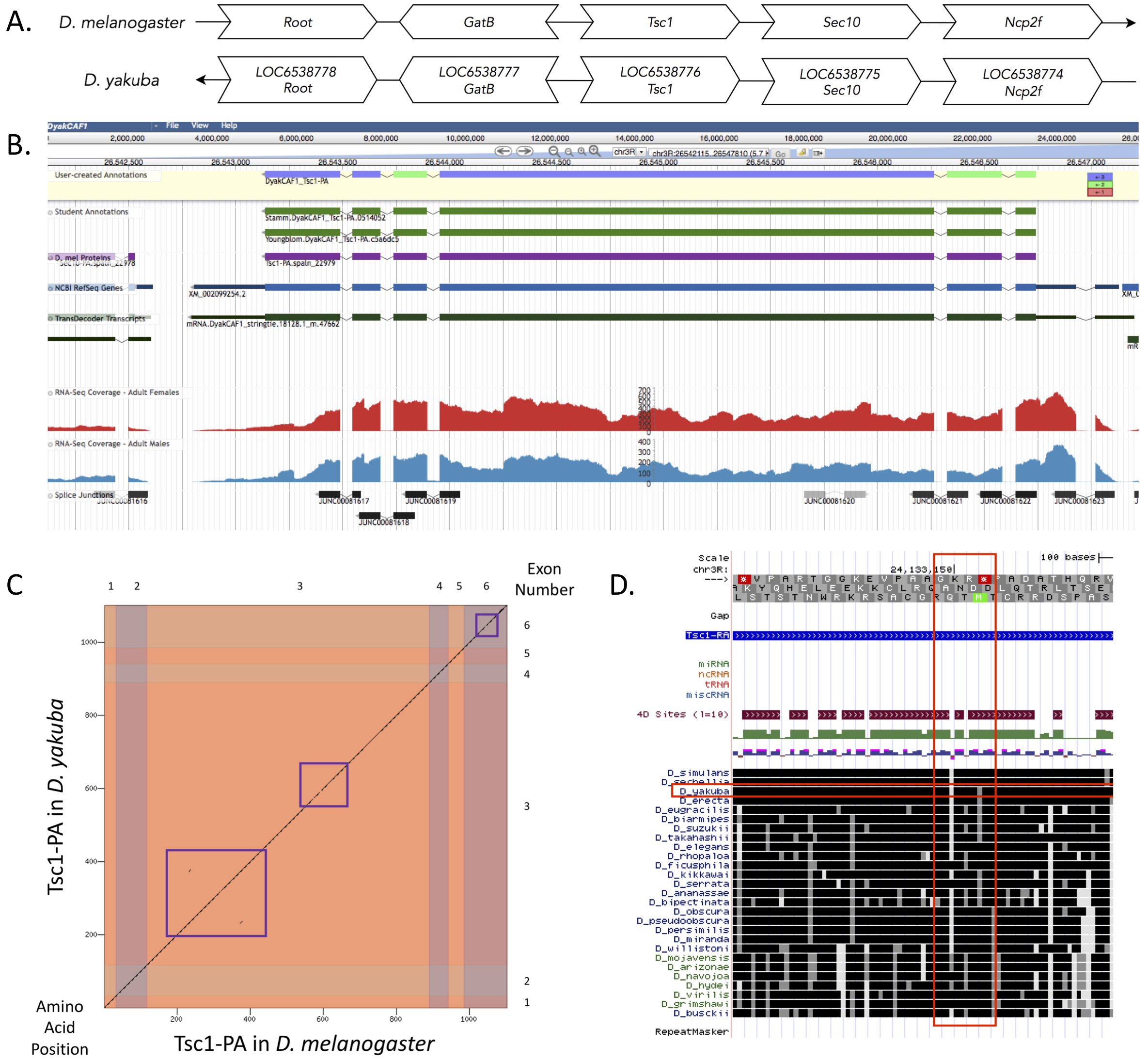
Gene Model for the ortholog of Tsc1 in the Drosophila yakuba DyakCAF1 assembly (GCA_000005975.1).
Description
Introduction
Tsc1 (LOC6538776) in D. yakuba is an ortholog to the Tsc1 gene in D. melanogaster. We used the D. yakuba CAF1 assembly (GCA_000005975.1, Drosophila 12 Genomes Consortium et al., 2007) and the D. melanogaster dm6 assembly (GCA_000001215.4, Release 6.32 FB2021_01). Mutations in either the Tsc1 or Tsc2 gene can cause the hamartoma syndrome tuberous sclerosis complex (TSC) (Dabora et. al, 2008). These two genes operate together in the insulin signaling pathway as tumor suppressors because of their ability to control cell growth (Gao, 1970). A mutation in the Tsc1 gene can also cause benign tumors to form in multiple organs (Potter, Huang, Xu, 2001). The NCBI RefSeq predicted model in D. yakuba, with a RefSeq accession number of XM_002099254.2 (RefSeq Release 204), has the same number of exons as the Tsc1 gene (LOC6538776) in D. melanogaster indicating they have an orthologous relationship. The methods and dataset versions used to establish the gene model are described in Rele et al. (2021). The GEP maintains a mirror of the UCSC Genome Browser (Kent WJ et al., 2002; Gonzalez et al., 2020), which is available at https://gander.wustl.edu and contains additional information about data sources and versions.
Synteny
The Tsc1 gene, located on chromosome 3R in D. melanogaster, is neighboring the genes Root, GatB, Sec10, and Ncp2f. The best candidate for the Tsc1 ortholog gene in D. yakuba based on the tblastn search is found on chromosome 3R. The candidate is also surrounded by the genes LOC6538778, LOC6538777, LOC6538775, and LOC6538774 (which are likely orthologous to Root, GatB, Sec10, and Ncp2f in D. melanogaster respectively, Figure 1A). We performed a blastp search of protein sequence XP_002099290.1 in D. yakuba against the protein sequences found in the refseq_protein database for D. melanogaster and it showed a high percent identity to Tsc1 in comparison to the second-best hit. After confirming that the genes surrounding Tsc1 are orthologous between the two species and the blastp results indicated a high percent identity for the Tsc1 gene between the two species, we determined that this region contains the ortholog for Tsc1 in D. yakuba.
Gene Model
Tsc1 has one isoform in D. yakuba, Tsc1-PA, with six exons. There are also six exons in the Tsc1 gene located in D. melanogaster. A blastp search of the protein sequence of Tsc1 in D. yakuba against D. melanogaster yields only one significant match with a 97.00% identity with only 33 amino acids differing out of 770. There was a small lack of sequence similarity between the protein sequences of the two species in coding exons three and six as is displayed by the purple boxes in the dot plot (Figure 1C). The large lack of sequence similarity in exon six, shown by the red vertical box in Figure 1D, can also be seen in the conservation tracks of 28 different Drosophila species in the UCSC Genome Browser. The lack of sequence similarity in exon six is consistent with the lack of a functionally-characterized protein domain in that region of the gene (FB2021_04, released August 17, 2021). The coordinates of the curated gene models can be found in NCBI at GenBank/BankIt using the accession BK014573. These data are also available in Extended Data files below, which are archived in CaltechData.
Acknowledgments
We would like to thank Wilson Leung, who created and maintains the GEP technological infrastructure. We would also like to thank Rachael A. Cowan for helping us submit the microPublication. This publication is dedicated to the memory of Dr. James J. Youngblom.
Extended Data
Rele, C. P. 2021. Dataset: dyakCAF1_Tsc1-PA.pep (Version 1.0) [Data set]. CaltechDATA. https://doi.org/10.22002/D1.1994
Rele, C. P. 2021. Dataset:DyakCAF1_Tsc1.FNA (Version 1.0) [Data set]. CaltechDATA. https://doi.org/10.22002/D1.1993
Rele, C. P. 2021. Dataset: dyakCAF1_Tsc1-PA.gff (Version 1.0) [Data set]. CaltechDATA. https://doi.org/10.22002/D1.1995
References
Funding
This material is based upon work supported by the National Science Foundation under Grant No. IUSE-1915544 to LKR and the National Institute of General Medical Sciences of the National Institutes of Health Award R25GM130517 to LKR. The Genomics Education Partnership is fully financed by Federal moneys. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed By
AnonymousHistory
Received: February 19, 2021Revision received: September 15, 2021
Accepted: September 16, 2021
Published: November 12, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Lose, B; Myers, A; Fondse, S; Alberts, I; Stamm, J; Youngblom, JJ; Rele, CP; Reed, LK (2021). Drosophila yakuba – Tsc1. microPublication Biology. 10.17912/micropub.biology.000474.Download: RIS BibTeX