Abstract
The utilization of plant gamma-aminobutyric acid (GABA) is essential for the pathogenicity of the bacterial plant pathogen Ralstonia solanacearum. A knockout mutant in the GABA transaminase-encoding gene gabT is unable to utilize GABA as a nutrient and its ability to cause disease in plants is strongly compromised. However, the proximity of the gabD gene (encoding a succinate semialdehyde dehydrogenase) in the same operon raises the question of whether an impact on the gabD gene underlies or contributes to the virulence attenuation of the ΔgabT mutant. In this work, we use genetic complementation to show that the expression of the gabT gene is able to rescue the impaired virulence of the ΔgabT knockout mutant in tomato plants, confirming that the gabT-encoded GABA transaminase is indeed required for full virulence of R. solanacearum in a natural host plant.
Description
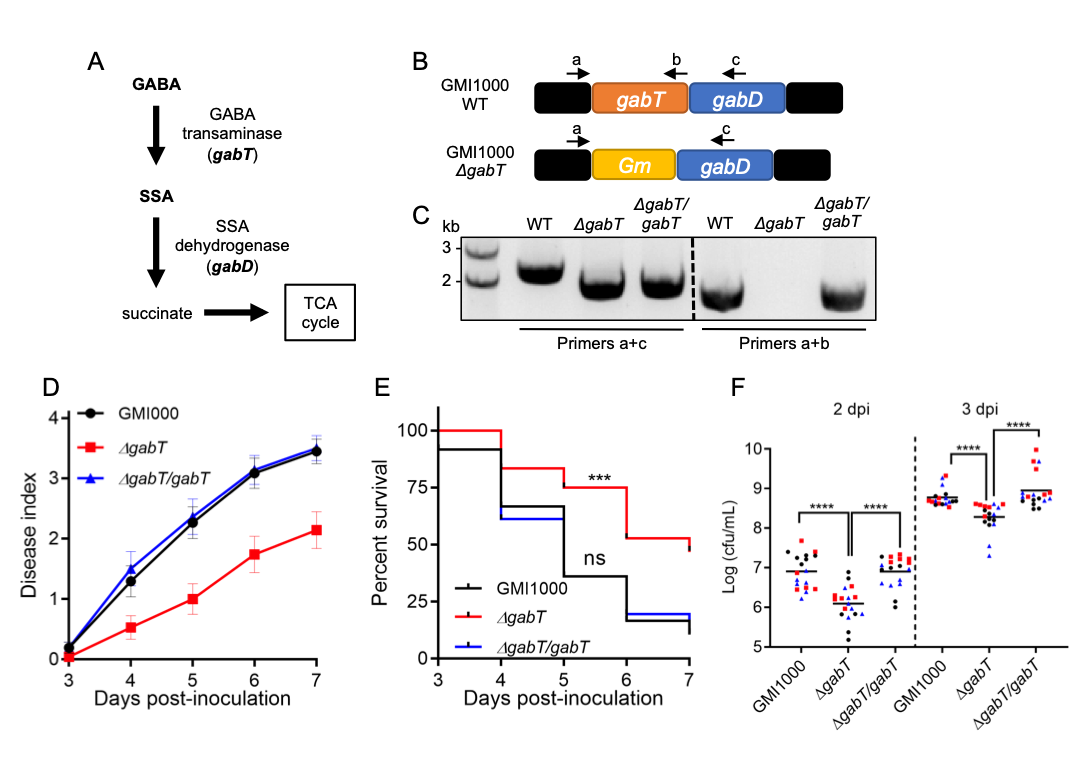
Ralstonia solanacearum is the causal agent of bacterial wilt disease of more than 250 plant species, and is considered as one of the most destructive plant pathogens (Mansfield et al., 2012). R. solanacearum is a soil-borne bacterium that enters plants through the roots, eventually reaching the vascular system; bacterial proliferation in xylem vessels causes vascular blockage, which leads to plant wilting and death (Xue et al., 2020). We have recently shown that R. solanacearum promotes the production of gamma-aminobutyric acid (GABA) in plant cells, which in turn can be used as a nutrient to support bacterial replication and pathogenesis inside the plant (Xian et al., 2020). In other gram-negative bacteria, such as R. eutropha (also known as Cupriavidus necator) or Pseudomonas syringae, GABA catabolism involves GABA transaminases (GabT), which convert GABA into succinate semialdehyde (SSA); SSA is used by succinate semialdehyde dehydrogenases (SSA-DH / GabD) to generate succinate, which can be incorporated into the TCA cycle (Lutke-Eversloh & Steinbuchel, 1999; Park et al., 2010; Figure 1A). The genome of R. solanacearum contains a single copy of the gabT gene (RSc0029), which is located upstream of the gabD gene (RSc0028) in the same operon (Figure 1B). Interestingly, a ΔgabT knockout mutant was unable to use GABA as a nutrient, and severely impaired in its ability to cause disease symptoms in the model plant Arabidopsis thaliana and to grow in stems of tomato plants, their natural host (Xian et al., 2020). These results suggest that R. solanacearum requires the GabT transaminase to cause disease. However, given that the gabT and gabD genes belong to the same operon, it is formally possible that the mutation in gabT affects the expression of gabD, which could also contribute to the reduced virulence observed in the ΔgabT mutant. In order to address this possibility, we complemented the ΔgabT mutant using an integrative plasmid that leads to the expression of the gabT gene driven by its own promoter (see methods; Figure 1C). First, we inoculated the ΔgabT mutant in tomato plants by soil-drenching, an inoculation method that mimics the natural infection route of R. solanacearum, and found that, as previously shown in Arabidopsis, the ΔgabT mutant is significantly impaired in its ability to cause disease symptoms in tomato, in comparison to the GMI1000 wild-type reference strain (Figure 1D and 1E). Importantly, the symptoms induced by the complementation strain expressing gabT in the ΔgabT mutant background (ΔgabT/gabT) were similar to those induced by the GMI1000 wild-type strain (Figure 1D and 1E). Moreover, upon injection in tomato stems, expression of the gabT gene was able to rescue the attenuation in growth observed in the ΔgabT mutant (Figure 1F). Altogether, these results indicate that the virulence attenuation observed in the ΔgabT mutant (Xian et al., 2020; Figure 1D and 1E) is indeed caused by mutation of the gabT gene. These results also suggest that either the gabD gene is still expressed in the ΔgabT mutant, or the gabD gene is not required for virulence. However, if the gabD gene is not required for virulence, how does R. solanacearum catabolize SSA to complete the GABA catabolic pathway? In R. eutropha it has been shown that a gabD mutant still displays SSA-DH activity (Lutke-Eversloh & Steinbuchel, 1999), suggesting the presence of additional SSA-DH enzymes, and such non-GabD SSA-DH enzymes have been identified in other bacteria, such as Escherichia coli or Klebsiella pneumoniae (Marek & Henson, 1988; Sanchez et al., 1989). Similarly, even if the gabD gene were affected in the ΔgabT mutant, it is possible that R. solanacearum uses other SSA-DH enzymes to complete the GABA catabolic pathway in the ΔgabT/gabT complementation strain.
Methods
Request a detailed protocolPlant material and growth conditions
Solanum lycopersicum
Tomato plants (Solanum lycopersicum cv. Moneymaker) were cultivated in jiffy pots (Jiffy International, Kristiansand, Norway) in a growth chamber under controlled conditions (25°C, 16 h light/8 h dark photoperiod, 130 mE m-2 s-1, 65% humidity) for 4 weeks. After soil drenching inoculation, the plants were kept in a growth chamber under the following conditions: 75% humidity, 12 h light, 130 mE m-2 s-1, 27°C, and 12 h darkness, 26°C for disease symptom scoring.
Bacterial strains
Ralstonia solanacearum strains, including the phylotype I reference strain GMI1000, GMI1000 ΔgabT mutant, and GMI1000 ΔgabT/gabT complementation strains were cultured overnight at 28 °C in complete BG liquid medium (Plener et al., 2010).
Generation of R. solanacearum gabT complementation strain
To complement gabT in the ΔgabT mutant (Xian et al., 2020), the gabT gene (RSc0029) with its promoter (300 bp upstream of ATG of the gabT gene) was amplified by PCR, cloned into pENTR-TOPO, and then introduced into pRCT-GWY (Monteiro et al., 2012) by LR reaction (in vitro recombination between the entry clone – pENTR-TOPO- and the destination vector – pRCT-GWY), resulting in the pRCT-pgabT–gabT (Vailleau et al., 2007) plasmid. The integrative pRCT-pgabT–gabT plasmid, triggering the expression of gabT under the control of the native gabT promoter, was transformed into the R. solanacearum ΔgabT mutant strain (in GMI1000 background) by natural transformation, leading to the stable integration of the pgabT-gabT fragment into a permissive site of the bacterial chromosome, as previously described (Monteiro et al., 2012). The complementation strain ΔgabT/gabT was selected using Tetracycline (10 μg mL-1) and confirmed by PCR using gabT specific primers (Figure 1b and Table S1).
Pathogen inoculation assays
For R. solanacearum soil-drenching inoculation, 12 3-4 week-old tomato plants for each bacterial strain (grown in Jiffy pots) were inoculated by soil drenching with a bacterial suspension containing 108 colony-forming units per mL (CFU mL-1). 300 mL of inoculum of each strain was used to soak each set of plants. After 20-minute incubation with the bacterial inoculum, plants were transferred from the bacterial solution to a bed of potting mixture soil in a new tray (Vailleau et al., 2007). Scoring of visual disease symptoms on the basis of a scale ranging from ‘0’ (no symptoms) to ‘4’ (complete wilting) was performed as previously described (Vailleau et al., 2007).
Stem injection assays with R. solanacearum were performed as previously described (Yu et al., 2020; Wang et al., 2021a; Wang et al., 2021b). Briefly, 5 μL of a 106 CFU mL-1 bacterial suspension were injected into the stems of 4-week-old tomato plants and 2.5 μL xylem sap were collected from each plant for bacterial quantification 2 or 3 days-post inoculation (dpi). Injections were performed 2 cm below the cotyledon emerging site in the stem, while the samples were taken at the cotyledon emerging site. CFUs were counted by spreading serial dilutions on solid BG medium (Plener et al., 2010).
Acknowledgments
We thank Xinyu Jian for technical and administrative assistance during this work, Rosa Lozano-Duran for critical reading of this manuscript, and all the members of the Macho laboratory for helpful discussions.
References
Funding
Work in the Macho laboratory is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB27040204), the Chinese 1000 Talents Program, and the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences).
Reviewed By
Caitilyn AllenHistory
Received: May 31, 2021Revision received: September 21, 2021
Accepted: September 24, 2021
Published: September 29, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Xian, L; Yu, G; Macho, AP (2021). The GABA transaminase GabT is required for full virulence of Ralstonia solanacearum in tomato. microPublication Biology. 10.17912/micropub.biology.000478.Download: RIS BibTeX