Abstract
The amino acid sequence Arg-Gly-Asp (RGD) is a cell-binding motif for extracellular matrix proteins. Initially found in fibronectin, the RGD motif is also found in LAM-3/laminin α chain in C. elegans. Laminin, a heterotrimeric glycoprotein, is a significant component of the basement membrane. Mutations in laminin subunits disrupt the extracellular matrix hence inhibit cell adhesion. This study aims to characterize the function of the RGD motif in lam-3/laminin α. Two mutations, lam-3 RGE and lam-3 ΔRGD, were generated. Our analysis of the mutants revealed that the RGD motif is involved in the motility of animals, suggesting that the cell-laminin interaction plays a role in regulating body contraction.
Description
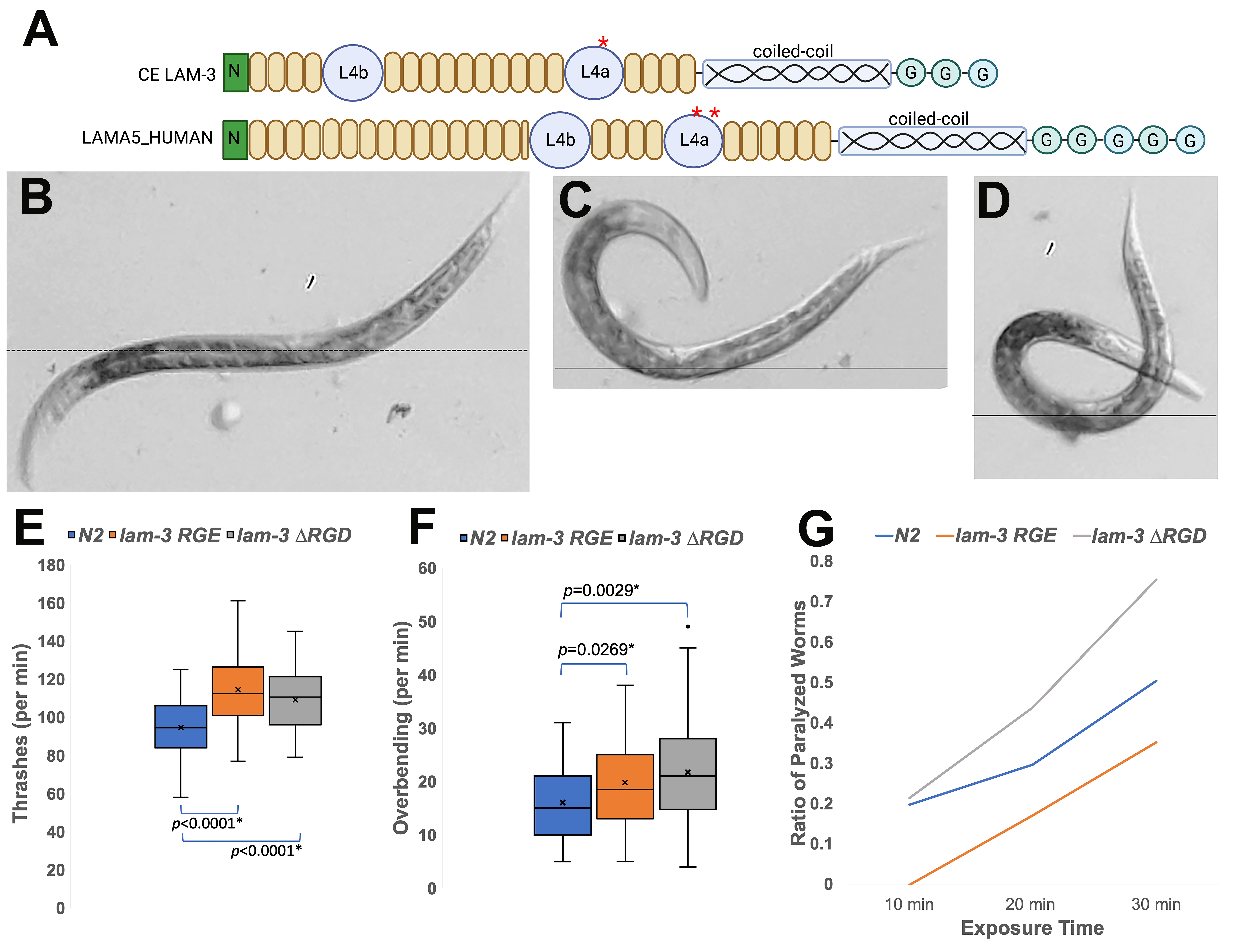
Laminin is a heterotrimeric glycoprotein composed of α, β, and γ subunits, one of the significant components in the basement membrane extracellular matrix (ECM). It plays an essential role in tissue organization such as ECM assembly, cell migration, and cell adhesion (Colognato and Yurchenco 2000). In the ECM, laminin interacts with cell surface receptors such as integrins and dystroglycan, linking ECM to actin cytoskeletons in muscle cells (Gawlik et al. 2006; Meinen et al. 2007). Among the laminin subunits, several laminin α chains bind to integrins. There are five α chains in humans. LAMA5 (laminin α5) contains the RGD motifs in its laminin IV type A domain (L4a). In C. elegans, LAM-3/laminin α possesses an RGD motif in the L4a domain, similar to the RGD location in LAMA5 (Figure 1A). Dystroglycan is a non-integrin binding receptor that bridges laminin to the cytoskeleton in skeletal muscle cells. Dystrophin and other transmembrane proteins are collectively identified as the dystrophin-glycoprotein complex that binds to laminins. Deficiencies in dystroglycan complex and laminin are linked to congenital muscular dystrophy (Sciandra et al. 2007; Sciandra et al. 2015). We surveyed ECM proteins and found an RGD (Arg-Gly-Asp) motif in LAM-3/laminin α (amino acid numbers 1462, 1463, and 1464). The RGD motif was edited to RGE or deleted to study the function of the motif.
In order to characterize the function of the RGD motif of LAM-3, the RGD motif was removed from the lam-3 locus by using the CRISPR-Cas9 system, which produced a mutant allele lam-3 (kq1461), designated as lam-3 ΔRGD. The RGD sequence is also changed to RGE (Arg-Gly-Glu), which produces another allele, lam-3 (kq1464), designated as lam-3 RGE (see Methods section). Previous studies on the ability of laminins to bind to integrins and dystroglycan (Yurchenco et al. 2018) hypothesized that mutations of the lam-3 RGD motif would result in locomotion anomalies. The thrashing (swimming) behavior in liquid medium has been a well-established and efficient method for measuring motility (Koopman et al. 2019). Thrashing is a coordinated contraction; the worm synchronously bends the anterior and posterior parts of body wall muscle cells. Typical thrashing behavior is characterized as worms swinging from one side to the other of the body axis (Figure 1B). Overbending movement occurs when muscle contraction is not harmonious across the body, generating a stop in either a “hook” position (Figure 1C) or an overbent U-shape (Figure 1D) (Ackley et al. 2003). When performing the thrashing assays, the number of overbending (Figures 1C and 1D) is not included in the thrashing counts. One-minute thrashes after a brief acclimation in M9 buffer showed that thrashing of both lam-3 RGE (114 thrashes per min, n=50) and lam-3 ΔRGD (109 thrashes per min., n=50) are significantly higher than that of N2 (95 thrashes per min. (n=50), p < 0.0001) (Figure 1E). The number of thrashes between lam-3 RGE and lam-3 ΔRGD was not statistically different (p = 0.2145). Then, we also measured the overbending. The result showed increased hyper-contraction in lam-3 RGD mutants, lam-3 RGE (20 overbends per min., n = 50) and lam-3 ΔRGD (22 overbends per min., n = 50), compared to N2 (16 overbends per min., n = 50, p < 0.05) (Figure 1F).
Bessou et al. found that mutants with muscle defects may show hypersensitive or resistant phenotypes to levamisole. This nicotinic drug targets levamisole-sensitive acetylcholine receptors (AChR) ion channels (Bessou et al. 1998). Levamisole stimulates the opening of the ion channels, promoting the entry of calcium ions, thus stimulating muscle contractions at neuromuscular junctions (Sloan et al. 2015). Levamisole response of the lam-3 mutants (Figure 1G) revealed that, after 30-minute exposure to 100 µM levamisole, 76% of lam-3 ΔRGD worms were paralyzed (n = 100), while the 35% of lam-3 RGE (n = 105) were paralyzed. This result suggested that lam-3 ΔRGD is hypersensitive to levamisole, while the lam-3 RGE mutant was resistant to the chemical.
To identify additional defects of lam-3 mutants, we also measured the touch sensitivity of the lam-3 mutants. C. elegans has six touch receptor neurons anchored closely to the cuticle and surrounded by epidermal cells. These touch neurons allow worms to detect and produce responses to external forces (Chalfie and Sulston 1981). Stroking at either the anterior or posterior part of a responsive worm with the tip of human hair elicits a type of avoidance behavior (Krieg et al. 2015). Our results showed that both lam-3 RGE (5%, n = 200, p > 0.011) and ΔRGD (5%, n = 200, p < 0.011) showed significant non-responsiveness to gentle touches compared to N2 (1.5%, n = 200), suggesting that the anchoring of touch neurons depends on the proper laminin function that ensures cell-matrix interaction.
We conclude that the mutations of lam-3 RGD result in increased body thrashing, hyper-contraction, and defective mechanosensation. Responses to levamisole in lam-3 RGD mutants confirm that lam-3 is vital for muscle contraction of the body. In C. elegans, hyper-contraction and levamisole resistance are typical phenotypes of dystrophic muscles (Chaya et al. 2021; Ellwood et al. 2021). Mutations in the dys-1/dystrophin gene display hyperactive muscle contraction and levamisole resistance due to defective cholinergic transmission across neuromuscular junctions (NMJ) (Bessou et al. 1998). The lam-3 RGD mutants may carry similar defects that lead to dystrophic muscle due to the defective NMJ. Future studies should further investigate the role of C. elegans ECM on the regulation of muscle functions.
Methods
Request a detailed protocolTo delete the RGD motif in lam-3 locus in chromosome I, we have identified an effective CRISPR site in the area of three amino acid numbers, 1462, 1463, and 1464, from the CRISPR guide RNA Selection Tool, http://genome.sfu.ca/crispr/search.html. According to the intended mutation, 94-mer repair DNA templates were designed and custom-made from IDT Inc., Coralville, IA, including all other PCR oligos in this study (Reagents section). Then, the mixture of template DNA (custom), crRNA (custom), tracr RNA (catalog no. 1072532), and Alt-R Cas9 (catalog no. 1081058) proteins are annealed at 37°C and micro-injected into the syncytial gonad arms of N2 animals (P0) with dpy-10 crRNA as a co-CRISPR marker (Paix et al. 2015; Dickinson and Goldstein 2016). The F1 offspring of P0 worms is selected by Dpy phenotype and is subjected to PCR genotyping to identify worms carrying the desired mutations. Once the F1 mutants are isolated, F2 progeny are screened with wild-type specific or mutant specific primer sets to identify the homozygote alleles. Isolated PCR products were also sequenced to confirm the mutations. A thrashing assay was performed with young adult worms.
The one-minute counting of thrashing is conducted after one minute of acclimation in 40 μl of M9 buffer. Videos for thrashing assays were taken using mobile devices. The counting of thrashing and overbending was performed manually. Continuous and simultaneous bending of both the head and the tail towards either side of the axis was counted as one thrash. Touch sensitivity assays (gentle touch) were performed by stroking at the posterior to the pharynx and near the tip of the tails (Goodman and Schwarz 2003). The elicited response characterized as ‘responsiveness’ is a changing of movement from forward to backward. Non-responsiveness is the lack of characteristic responsiveness after stroking at both ends. A worm pick with fine hair attached to the end was used for the stroke. To characterize levamisole phenotypes, we performed a levamisole sensitivity assay by placing worms in 100 µM levamisole in M9 solution for thirty minutes and counting the number of paralyzed worms in ten-minute intervals. The paralysis was tested by a gentle stirring, the same tool aforementioned in the touch assay. The lack of muscle contraction in response to a mechanical stimulus is regarded as paralysis. All statistical analyses in this study were performed with Wilcoxon Analysis (Preisser et al. 2011) using JMP Pro 15.2.0 (SAS lab, Cary, PA).
Reagents
DNA template sequence for homology directed repair
| Temp-LAM3RGD1464D | CAAGACACCCAAGGAATATATACCGTGGAACCTATACTTACCCGGCTGCAA TTAACATCCAAGAGGTTTCCCTTGACGTAGCTGTTCCTGAATC |
| Temp-LAM3RGE1464 | CAAGACACCCAAGGAATATATATCCGTGGAACCTATACTTACCCGGCTCGA GGCGAAGCTATCAATATTCAAGAGGTTTCCCTTGACGTAGC |
crRNA for lam-3 locus
| LAM3RGD | ACCTACACATATCCAGCAAG | AGG (PAM sequence) |
co-CRISPR dpy-10 crRNA
ZQDP10A: GCTACCATAGGCACCACGAG
Genotyping and Sequencing Primers
| LAM3GRESEQF | GTCACTCTCCAGAGCTCACAC |
| LAM3RGESEQR | CAGTGCTCACAGAAATCACCG |
| LAM3RGDWTF | CTACACATATCCAGCAAGAGGTGAT |
| LAM3RGEF | CCGGCTCGAGGCGAA |
| LAM3RGD1464DF | TACTTACCCGGCTGCAATTAACATC |
| Strain | Genotype | Available from |
| N2 | Wild Type Caenorhabditis elegans Bristol Strain | CGC |
| BU1461 | lam-3 (kq1461) | This study |
| BU1464 | lam-3 (kq1464) | This study |
Acknowledgments
The authors acknowledge BIO4108 Cell and Developmental Biology lab students (2019 – 2020) who participated in the LAM-3 RGD characterization project. We give special thanks to Meg Taylor, Jess Root, and Kyle Dinh for their contributions during the initial stage of LAM-3 projects.
References
Funding
Funding for this project was provided by Baylor University.
Reviewed By
AnonymousHistory
Received: September 11, 2021Revision received: September 27, 2021
Accepted: September 27, 2021
Published: October 11, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Wang, L; Qiu, Z; Lee, M (2021). Mutations in the cell-binding motif of lam-3/laminin α reveal hypercontraction behavior and defective sensitivity to levamisole in Caenorhabditis elegans. microPublication Biology. 10.17912/micropub.biology.000485.Download: RIS BibTeX