School of Biosciences, University of Melbourne, Parkville, Victoria 3010, Australia.
Abstract
Arabidopsis thaliana MYB5 collaborates with TRANSPARENT TESTA GLABRA1 (TTG1) and basic-Helix-Loop-Helix (bHLH) transcription factors to regulate seed coat, trichome and root cell differentiation. Using a yeast two-hybrid system we show that the N-terminal region of MYB5 binds directly to the serine/threonine CASEIN KINASE2 BETA3 (CK2β3) subunit. Functions of the CASEIN KINASE2 (CK2) complex include facilitating phosphorylation of MYB transcription factors and cell cycle checkpoint regulatory proteins. Purified recombinant MYB5 protein was found to bind only weakly in vitro to the promoter of ALPHA/BETA ESTERASE/HYDROLASE4 (ABE4), a known MYB5 target gene. We propose that phosphorylation of MYB5 facilitated by the MYB5-CK2β3 interaction enhances MYB5 binding to its target gene promoters.
Description
The Arabidopsis (Arabidopsis thaliana) MYB5 (At3g13540) transcription factor (TF) forms transcriptional regulatory (MBW) complexes with basic-Helix-Loop-Helix (bHLH) and TRANSPARENT TESTA GLABRA1 (TTG1) (WD-repeat) TF proteins. These MBW complexes regulate seed coat, trichome and root cell differentiation via a multi-tiered transcriptional mechanism (Golz et al., 2018; Li et al., 2020). MYB5 is partially redundant with MYB23 and TRANSPARENT TESTA2 (TT2) (another MYB protein, MYB123) in MBW complex formation. TTG1 is subject to site-specific phosphorylation by SHAGGY-LIKE KINASE11 (SK11) and SK12 which inhibits TTG1 interactions with TT2 (Li et al., 2018). MYB5 also possesses potential phosphorylation sites at the N-terminus (Li et al., 1996).
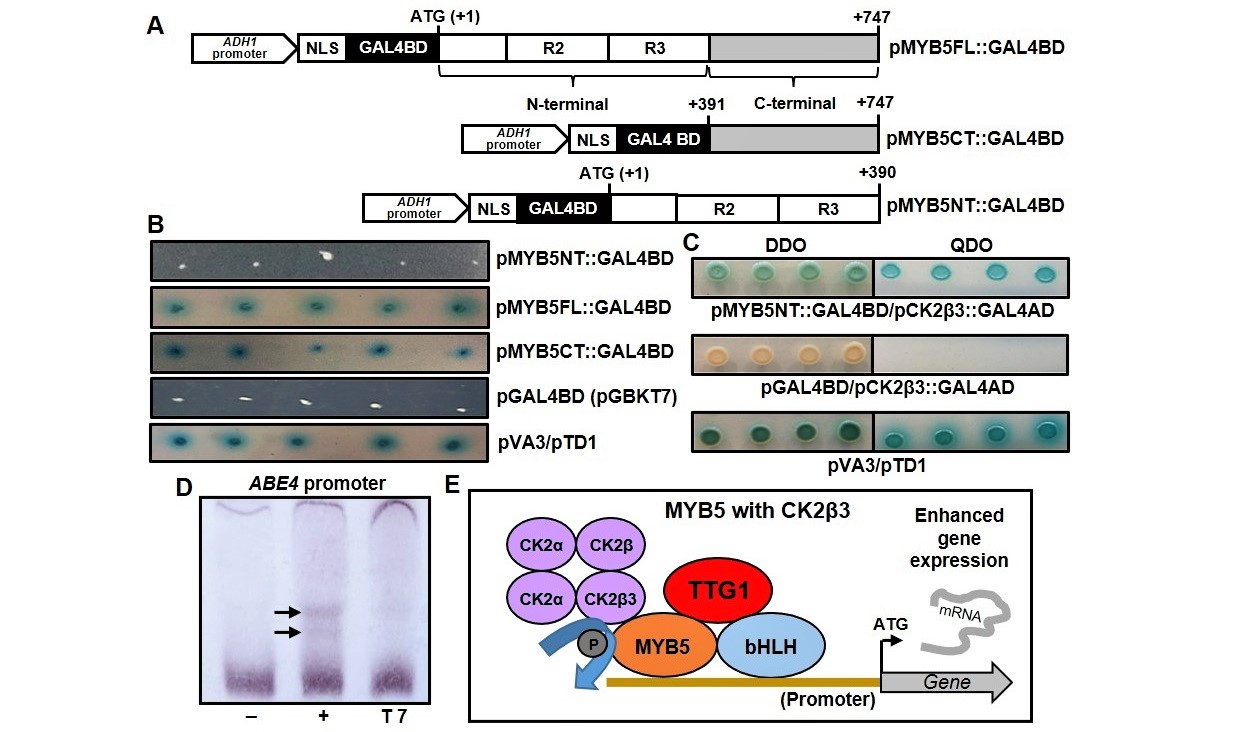
To identify proteins interacting with MYB5, the Matchmaker® Gold Yeast Two-hybrid system was used to screen a prey cDNA library. Three candidate bait constructs contained cDNA nucleotide sequences encoding the MYB5 full length, C-terminal and N-terminal regions (Fig 1A), respectively. The constructs containing full length MYB5 and the MYB5 C-terminal region activated MEL1 and AUR1-C reporter genes resulting in blue colonies (Fig 1B) indicating that the C-terminal region acts as an activator motif in yeast. The MYB5 N-terminal construct (pMYB5NT::GAL4BD) failed to activate the reporter genes (Fig 1B) and was therefore used as the bait for library screening. On the screening plates, five yeast colonies were identified and their prey cDNA fragments sequenced. Three of the five cDNA sequences encoded a serine/threonine protein kinase CAESIN KINASE2 BETA3 (CK2β3) (At3g60250) subunit fused in frame with the GAL4AD domain-encoding nucleotide sequence. The remaining two cDNA sequences encoded Hypothetical/HEAT RESPONSE PROTEIN (At5g10010) and PHOSPHOGLUCOSE ISOMERASE (At4g24620). The dependency of pCK2β3::GAL4AD on pMYB5NT::GAL4BD in activating four reporter genes, namely AUR1-C, HIS3, ADE2 and MEL1, was then confirmed (Fig 1C).
MYB5 directly regulates ALPHA/BETA ESTERASE/HYDROLASE1 (ABE1, At2g23550) and ABE4 (At2g23580) expression (Li et al., 2020). The electrophoretic mobility shift assay (EMSA) was used to determine in vitro binding of purified MYB5 protein to the ABE4 promoter. A truncated MYB5 protein containing the MYB DNA-binding domain was expressed in E. coli and purified. The ABE4 promoter probe (Suppl. Fig 1) contains two sequences identified as MYB5 binding sites using chromatin immunoprecipitation (Li et al., 2020). The EMSA identified two weak band shifts (Fig 1D) indicating that MYB5 binds weakly to the ABE4 promoter region in vitro and may therefore be subject to phosphorylation/activation for enhanced MYB-DNA binding in vivo.
The Arabidopsis CK2β3 subunit is part of the CK2 holoenzyme which is a tetramer consisting of two regulatory β-subunits and two catalytic α-subunits collectively implicated in a number of developmental and stress-responsive pathways (Mulekar and Huq, 2013; Wei et al., 2021). Known CK2 substrates primarily include transcription factors or regulatory proteins (Mulekar and Huq, 2013). CK2β3 is highly expressed in developing seeds and has an expression pattern overlapping that of MYB5, TTG1, ABE1 and ABE4 (Winter et al., 2007). CK2β3 localises to the nucleus and cytosol (Salinas et al., 2006) and can modulate the activity of the MYB-related CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) both by direct physical interaction in vivo and phosphorylation (Sugano et al., 1999). CK2β3 shares 75% amino acid sequence similarity with CK2β1 and 71% with CK2β2 (Sugano et al., 1998). CK2β3 and CK2β4 subunits share 87% amino acid sequence identity and 92% sequence similarity. Moreover, like CK2β3, CK2β4 has also been linked to the phosphorylation of CCA1 as over-expression of both genes leads to period-shortening of circadian clock-related gene expression (Perales et al., 2006). Although CK2β3 does not exhibit phosphorylation activity, it has been shown to facilitate CCA1 binding to the promoter of target gene CHLOROPHYLL A/B BINDING PROTEIN1 (CAB1) (Sugano et al., 1998). CK2 also interacts with the MYB-related LATE ELONGATED HYPOCOTYL (LHY) and YING YANG1 (YY1) via CK2β3 and phosphorylates LHY and YY1 proteins in vitro (Sugano et al., 1999; Wu and Li, 2017).
The transcriptional activities of Pinus taeda MYB4 (PtMYB4) and PtMYB46 are positively regulated by phosphorylation (Morse et al., 2009) although phosphorylation of TFs such as LTF1 (which regulates lignin biosynthesis in Populus) can result in its degradation (Gui et al., 2019). In contrast, DNA binding of Arabidopsis MYB15, a transcriptional repressor of cold signalling, is reduced by phosphorylation (Kim et al., 2017). While specific MYB5-CK2β3 interactions in planta are yet to be determined, CK2β3 may facilitate direct MYB5 binding to its target gene promoters or may phosphorylate MYB5 indirectly by recruiting the CK2 catalytic α-subunits. The MYB5 N-terminal region contains several threonine and serine residues which are potential phosphorylation sites (Li et al., 1996) and phosphorylation of these residues may enhance MYB5 protein-DNA binding affinity. Alternatively, as MYB5-TTG1 and/or MYB5-bHLH interactions were not detected in the initial yeast two-hybrid screening, MYB5 may also require activation to facilitate TF protein-protein binding. TTG1 is subject to site-specific phosphorylation which regulates binding with TT2 (MYB123) to form some MBW complex combinations (Li et al., 2018). MYB5-CK2B3 interactions and proposed phosphorylation may contribute to MYB5-bHLH-TTG1 (MBW) complex formation which, in turn, may further increase MYB5 protein-DNA binding affinity in vivo.
In the proposed model (Fig 1E), MYB5-bHLH-TTG1 complexes activate the expression of target genes by binding to their promoters. CK2β3 interacts with MYB5 transiently and recruits the CK2 holoenzyme to phosphorylate MYB5 (Fig 1E) resulting in more active MYB5-bHLH-TTG1 complexes thereby enhancing expression of MYB5 target genes.
Methods
Request a detailed protocolPlasmid construction
For yeast two-hybrid (Y2H) analysis, MYB5 full length, C-terminal and N-terminal nucleotide sequences were PCR amplified from cDNA prepared from developing wild-type (Col) siliques using Y2HFULLF/Y2FULLR [pMYB5FL], Y2HFULLF/Y2HNTR [pMYB5CT] and Y2HCTF/Y2FULLR [pMYB5NT] primer combinations, respectively. The MYB5 cDNA sequences were ligated into the pGBKT7 yeast expression vector at the EcoR1 and BamH1 restriction sites, generating pMYB5FL::GAL4BD, pMYB5CT::GAL4BD and pMYB5NT::GAL4BD vectors, respectively. For EMSA analysis, a truncated MYB5-encoding cDNA sequence (410 nucleotides) containing the MYB DNA-binding domain sequence with the truncation after the GIDPOTHK polyadenylation site was PCR amplified from wild-type (Col) seed cDNA using MYBPROTF and MYBTRUNCR primers. Truncation of MYB proteins beyond the MYB domain does not appear to affect binding specificity (Li and Parish, 1995; Phan et al., 2011). The truncated MYB5 fragment was ligated into the pRSETA expression vector (Invitrogen) at Xho1 and EcoR1 restriction sites and was fused to a 6X polyhistidine tag and a T7 leader peptide coding sequence and driven by the T7 promoter which was PCR amplified using T7F and PGADT7R primers.
Yeast Two-Hybrid analysis
The yeast two-hybrid (Y2H) assay was performed using the Matchmaker™ Gold Yeast Two-Hybrid system (Clontech) and all protocols were adapted from the Clontech Yeast Protocols Handbook and the Matchmaker™ Gold Yeast Two-Hybrid user manual. To determine the autoactivation of candidate bait constructs, Y2HGold cells were transformed with the three constructs and inoculated onto dropout (-trp) media containing x-α-gal and auribasidin A. Five technical replicates are presented (Fig 1B). pGAL4BD (empty pGBKT7 vector) was used as a negative control and pVA3/pTD1 co-transformed cells were used as a positive control. The Mate & Plate™ Library – Universal Arabidopsis (Normalised) (Clontech) was used which consists of a cDNA library cloned into the GAL4AD vector (pGADT7, prey) and transformed into yeast strain Y187. Y2HGold yeast cells transformed with pMYB5NT::GAL4BD (bait) were mated with the Y187 cells expressing the Mate & Plate™ Library made from 11 Arabidopsis tissues. The mated cultures were plated onto the double dropout (–trp –leu) media (DDO) containing x-α-gal and auribasidin A. to identify bait and prey interaction. Yeast cells were then inoculated on double-dropout (DDO) medium (-leu/-trip) plus x-α-gal and quadruple-dropout (QDO) (-ade/-his/-leu/-trip) medium plus x-α-gal and auribasidin A. Double-dropout medium (-leu/-trp) selects for the presence of both pGADT7 and pGBKT7. Figure 1C depicts one representative biological replicate of MYB5-CK2β3 binding. Four technical replicates are presented. In total, 3 out of 5 (60%) of the positive colonies from initial Y2H screening were confirmed as positive MYB5-CK2β3 interactions.
Electrophoretic Mobility Shift Assay (EMSA)
Recombinant MYB5 protein expressed in E. coli was purified using Ni-NTA Superflow columns (Qiagen) according to the manufacturer’s protocol. EMSA was performed using a non-radioactive protocol (adapted from Phan et al., 2011) and the protocol described by Li and Parish (1995). The ABE4MCOREF and ABE4MCORER primers were designed to PCR amplify a probe of 150 to 200 nucleotides in length based on the promoter region enriched in ChIP analysis (Li et al., 2020). Protein-probe binding was performed using 400 ng total protein, 3 mL of binding buffer (20 mM NaCl, 5 mM MgCl2, 20 mM Tris [pH 8.0], 10% glycerol, 0.5 mM EDTA and 0.5 mM DTT) with 1 mL of poly d(I-C). The truncated MYB5 protein was expressed in E. coli and purified. SDS-PAGE and Coomassie Blue staining detected bands of approximately ~21kDa molecular weight in two independent replicates. Western Blot analysis using an anti-T7 epitope tag antibody detected ~21k kDA bands in each lane corresponding to the truncated MYB5 protein replicates. EMSA probes were labelled with digoxigenin via PCR using digoxigenin-labelled dUTP (Roche).
Reagents
Primer oligonucleotide sequences
| Primer name | Primer sequence (5′ to 3′ orientation) |
| Y2HFULLF | GAGAGAGAATTCATGATGTCATGTGGTGGG |
| Y2FULLR | GAGAGAGGATCCCTAGTCATGTCCTAAGCTAGAAGA |
| Y2HCTF | AGAGAGAATTCGGAATTGATCCTCAAACCCACAAG |
| Y2HNTR | GAGAGAGGATCCTTGCCTTAAAAGTTTCTTACGAAG |
| T7F | TAATACGACTCACTATAGGGC |
| PGADT7R | AGATGGTGCACGATGCACAG |
| MYB5PROTF | CGAGCTCGAGATGATGTCATGTGGTGGGAAGAAGCC |
| MYB5PROTR | GAGAGAATTCCTAGTCATGTCCTAAGCTAGAAGA |
| MYB5TRUNCR | TCTCGAATTCCTACTTGTGGGTTTGAGGATCAATTCC |
| ABE4MCOREF | GGTTCAGAATTTATTACTTACTTTGGTTG |
| ABE4MCORER | CGATGGTCACTTTCCTCATACTCTTTC |
Acknowledgments
We thank the current and former members of the Roger Parish lab (La Trobe University, Melbourne, Australia) for their generous assistance with yeast two-hybrid and EMSA experiments.
References
Funding
This work was supported in part by the Grains Research and Development Corporation (GRDC) (ACT, Australia), Australian Research Council (ARC) and Australian Government Research Training Program (RTP) Scholarships awarded to RSN and PJA.
Reviewed By
Vy NguyenHistory
Received: May 20, 2021Revision received: September 9, 2021
Accepted: September 29, 2021
Published: October 7, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Napoli, RS; Allen, PJ; Parish, RW; Li, SF (2021). The Arabidopsis MYB5 transcription factor interacts with CASEIN KINASE2 BETA3 subunit in a yeast two-hybrid system. microPublication Biology. 10.17912/micropub.biology.000486.Download: RIS BibTeX